 PDF(26977 KB)
PDF(26977 KB)


 PDF(26977 KB)
PDF(26977 KB)
 PDF(26977 KB)
PDF(26977 KB)
DNA中醛基嘧啶的化学检测
 ({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}
({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}Detection of 5-Formylpyrimidines in DNA Based on Chemoselective Labeling
 ({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}
({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}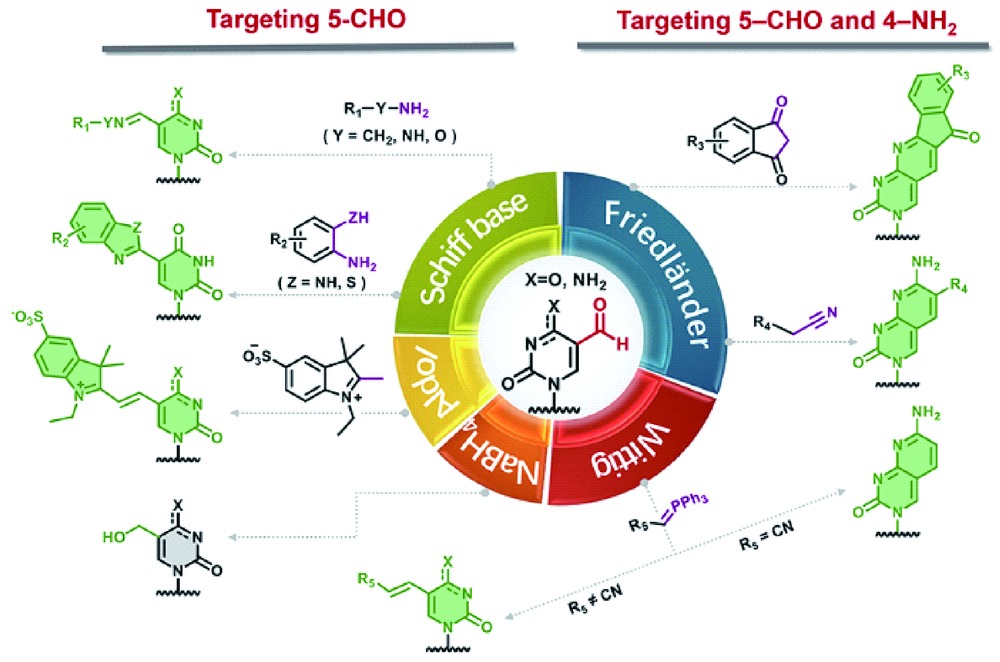
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
/
| 〈 |
|
〉 |