 PDF(14991 KB)
PDF(14991 KB)


 PDF(14991 KB)
PDF(14991 KB)
 PDF(14991 KB)
PDF(14991 KB)
基于微流控芯片的体外三维肝脏生理模型的构建及应用
 ({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}
({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}Construction and Application of 3D Microfluidic Liver-On-A-Chip
 ({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}
({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}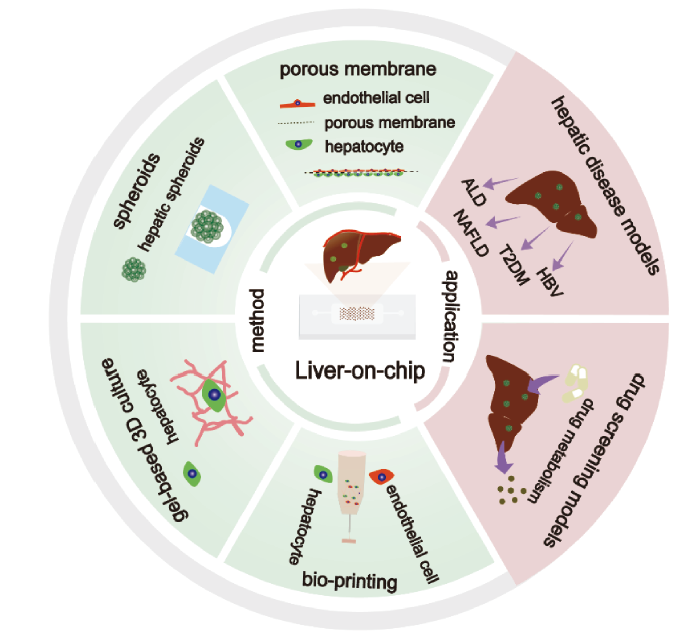
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
/
| 〈 |
|
〉 |