 PDF(1033 KB)
PDF(1033 KB)


 PDF(1033 KB)
PDF(1033 KB)
 PDF(1033 KB)
PDF(1033 KB)
生物大分子液-液相分离研究方法
 ({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}
({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}Research Methods for Liquid-Liquid Phase Separation of Biological Macromolecules
 ({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}
({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}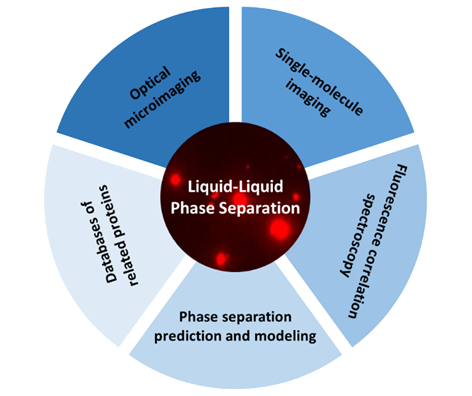
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
/
| 〈 |
|
〉 |