1 引言
2019年末,由一种新型冠状病毒感染的肺炎疫情(简称新冠肺炎,COVID-19)在世界多地、多点出现。2020年1月20日疫情在中国暴发之时,中共中央总书记、国家主席习近平对新冠肺炎疫情做出重要指示,强调要及时深化国际合作。抗击疫情期间,中国与美国、德国、日本、韩国、尼日利亚、俄罗斯、新加坡和世界卫生组织25名专家组成的中国-世界卫生组织联合专家考察组,与全球共享新冠病毒基因组信息,不断与各国疾控中心进行信息沟通和技术合作[1]。2020年8月24日,全球确诊病例已超过2 300万,与此同时,全球的科研机构争分夺秒开展攻关,在疫情期间独立或合作发表的关于新冠肺炎的科研成果数量快速增加。因此,有必要对相关研究进展进行总结梳理,并针对其中的国际合作情况开展分析。本研究使用文献计量法对此阶段的科研成果及国际合作进行了定量分析,利用VOSviewer工具进行了可视化展示,通过总结当前的科研成果和研究热点,了解国际合作的现状,可为该领域下一步的研究及政策制定提供建议和依据。
2 数据来源与分析方法
2.1 数据来源
选取WoS核心集数据库作为数据来源,检索策略基于新冠病毒及感染肺炎的命名过程制定。2020年1月7日,中国疾病预防控制中心从患者样本中鉴定出这种新型冠状病毒,随后世界卫生组织将其命名为“2019-nCoV”,后因其临床症状与严重急性呼吸综合征(SARS)非常相似,国际病毒分类委员会正式将其命名为“SARS-CoV-2”。2020年2月11日,世界卫生组织正式将这种病毒感染的疾病命名为“COVID-19”。因此本研究确定最终的检索式为:“TS=(2019-nCoV OR ‘2019 novel coronavirus’ OR SARS-CoV-2 OR COVID-19 OR Wuhan NEAR1 (coronavirus OR pneumonia))”,检索时间范围为2019年12月1日至2020年8月25日,检索日期为2020年8月26日,选择论文类型为Article、Early Access和Letter,检索出16 308篇,去重后共16 064篇。
2.2 数据分析方法
VOSviewer是众多科学知识图谱软件之一,可通过“网络数据”(文献知识单元)的关系构建和可视化分析,实现科学知识图谱的绘制,展现知识领域的结构、进化、合作等关系,其突出特点是图形展示能力强,适合大规模数据[2]。
首先利用VOSviewer对新冠肺炎相关研究主体的合作行为进行分析,包括国家/地区和机构的合作网络分析,可视化展示合作密集的区域,获取合作密切的国家/地区和机构。其次,重点对新冠肺炎相关研究进行主题聚类,绘制聚类图谱以归纳主要的研究主题,并具体分析每个主题下的代表性论文,同时具体分析合作紧密的中国与美国、英国之间的研究主题,比较中美、中英之间的科研差异。
3 结果与分析
3.1 合作网络分析
VOSviewer软件可进行机构、国家/地区的合作网络分析。合作网络图谱中,节点的大小表示发文量,节点越大代表发文量越大。节点间的连线表明两者的合作关系,线条越粗,表明合作的次数越多、合作越紧密、连接强度越高。节点的中心度是指所在网络通过该节点的任意最短路径的条数,代表节点在整体网络中所起的连接作用的大小,整个网络范围内的合作密切程度也可以用中心度衡量,合作网络图中紫色节点代表中心性较高,表示合作关系较紧密。
3.1.1 机构合作分析
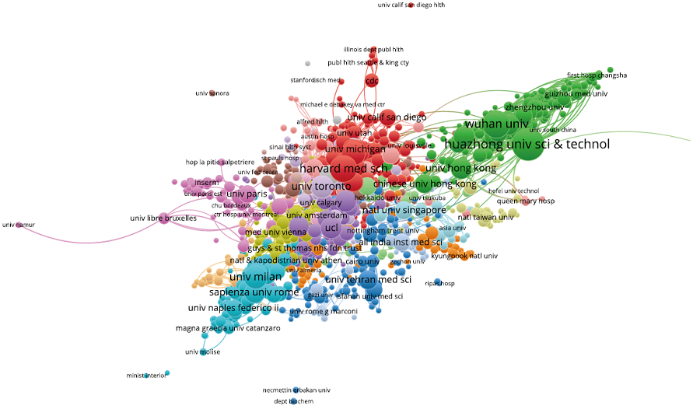
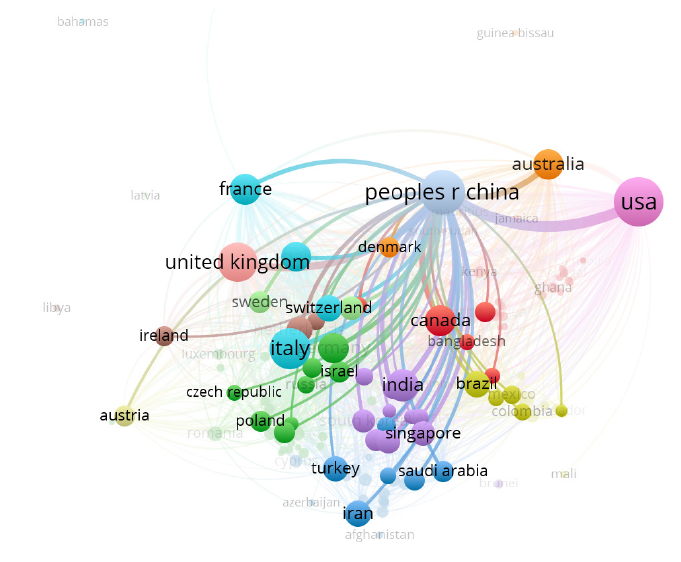
将新冠肺炎的相关文献数据导入到VOSviewer中,分析类型选择Co-authorship合著研究,分析单元选择Organizations,设置阈值为5,利用VOSviewer初步筛选出发文量≥5的1 682个发文机构。虽然VOSviewer的内置功能已初步对机构名称进行了统一大小写的去重工作,但对于作者著录时采用不同方式表述的同一机构名称仍需要进行清洗。首先,使用Google开发的开源数据清洗工具OpenRefine对机构名称进行自动聚类,然后,根据聚类结果人工判读是否为同一机构,最终获取规范化后的机构1 613个。最后将规范后的机构数据再次导入VOSviewer,绘制发文量≥5的1 613个研究机构的合作图谱(图1)和具体发文量排名表(表1)。
图1
表1 新冠肺炎发文机构Top30
| 排名 | 机构名 | 发文量/篇 | 排名 | 机构名 | 发文量/篇 |
|---|---|---|---|---|---|
| 1 | 华中科技大学 | 457 | 16 | 伦敦国王学院 | 132 |
| 2 | 武汉大学 | 296 | 17 | 宾夕法尼亚大学 | 130 |
| 3 | 哈佛医学院 | 281 | 18 | 荷兰莱顿大学 | 127 |
| 4 | 华盛顿大学 | 217 | 19 | 上海交通大学 | 126 |
| 5 | 米兰大学 | 208 | 20 | 帕多瓦大学 | 126 |
| 6 | 多伦多大学 | 187 | 21 | 首都医科大学 | 123 |
| 7 | 牛津大学 | 163 | 22 | 德黑兰大学医学院 | 122 |
| 8 | 伦敦大学学院 | 156 | 23 | 墨尔本大学 | 117 |
| 9 | 复旦大学 | 154 | 24 | 密歇根大学 | 117 |
| 10 | 罗马大学 | 152 | 25 | 香港中文大学 | 111 |
| 11 | 香港大学 | 151 | 26 | 帝国理工学院 | 109 |
| 12 | 中国科学院大学 | 146 | 27 | 新加坡国立大学 | 108 |
| 13 | 浙江大学 | 141 | 28 | 约翰斯·霍普金斯大学 | 107 |
| 14 | 哥伦比亚大学 | 140 | 29 | 全印度医学科学研究所 | 103 |
| 15 | 斯坦福大学 | 135 | 30 | 巴黎大学 | 101 |
从图1可以发现,研究机构主要分为两大部分:第一部分是发文较多、合作较紧密的位于中间区域的机构,第二部分是少数分散在外部区域的机构,其相互之间关联较少,说明合作比较稀疏。总体而言,大部分机构之间合作紧密,据VOSviewer统计,1 613个发文机构中有1 603个彼此之间有连线,表明彼此间存在合作关系。
由表1可知,前一阶段新冠肺炎的研究集中在中国武汉的高校及一些中外知名高校、病毒研究所和医院。排名前2位的华中科技大学和武汉大学均为中国武汉的高校,发文量分别为457篇和296篇。哈佛医学院位居第3,发文281篇。其他发文较多的机构依次是华盛顿大学(217篇)、米兰大学(208篇)、多伦多大学(187篇)、牛津大学(163篇)、伦敦大学学院(156篇)、复旦大学(154篇)、罗马大学(152篇)、香港大学(151篇)、中国科学院大学(146篇)、浙江大学(141篇)、哥伦比亚大学(140篇)等。总体而言,发文机构都是(医学类)高校机构。表1体现了中国科研机构在参与新冠肺炎临床救治时高效的科研攻关能力和快速响应能力。
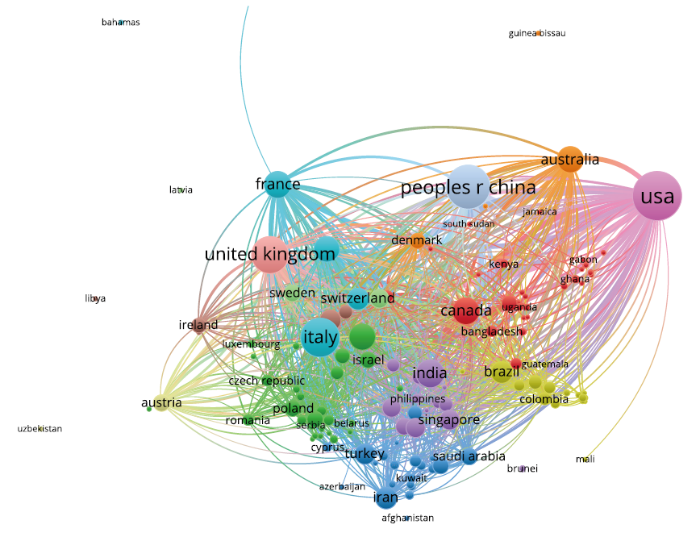
3.1.2 国家/地区合作分析
表2 新冠肺炎研究Top20发文国家/地区
| 序号 | 国家/地区 | 总发文量/篇 | 独立发文量/篇 | 独立发文量占比 | 总连接强度 |
|---|---|---|---|---|---|
| 1 | 美国 | 4374 | 2741 | 62.67% | 3592 |
| 2 | 中国 | 3099 | 2212 | 71.38% | 1776 |
| 3 | 意大利 | 2157 | 1462 | 67.78% | 2344 |
| 4 | 英国 | 1861 | 872 | 46.86% | 2906 |
| 5 | 印度 | 856 | 533 | 62.27% | 912 |
| 6 | 加拿大 | 761 | 307 | 40.34% | 1337 |
| 7 | 法国 | 747 | 364 | 48.73% | 1470 |
| 8 | 德国 | 692 | 280 | 40.46% | 1701 |
| 9 | 澳大利亚 | 667 | 252 | 37.78% | 1268 |
| 10 | 西班牙 | 623 | 312 | 50.08% | 1415 |
| 11 | 伊朗 | 406 | 269 | 66.26% | 428 |
| 12 | 巴西 | 393 | 242 | 61.58% | 642 |
| 13 | 瑞士 | 390 | 97 | 24.87% | 1159 |
| 14 | 荷兰 | 355 | 104 | 29.30% | 1176 |
| 15 | 土耳其 | 346 | 238 | 68.79% | 517 |
| 16 | 新加坡 | 301 | 173 | 57.48% | 486 |
| 17 | 日本 | 271 | 157 | 57.93% | 446 |
| 18 | 比利时 | 264 | 90 | 34.09% | 843 |
| 19 | 韩国 | 243 | 164 | 67.49% | 308 |
| 20 | 中国台湾 | 186 | 112 | 60.22% | 193 |
图2
图3
从发文数量看(表2),美国位居首位,共发表论文4 374篇,其中独立发文量为2 741篇。其次是中国,发表论文3 099篇,其中独立发表论文2 212篇。中、美明显领先其他国家,且中国独立发文量占比更高。意大利名列第3,发文2 157篇,其他发文较多的国家分别是英国(1 861篇)、印度(856篇)、加拿大(761篇)、法国(747篇)及德国(692篇)等。发文量的多少不仅和科研水平有关,还与该国是否大面积暴发疫情有一定关联。结合世界卫生组织在2020年8月25日公布的全球海外疫情数据,截至2020年8月24日下午14时,美国累计确诊病例数最多,发文量也位居世界第1。发文量位居第2的中国则是疫情暴发较早的地区。发文量较多的印度、西班牙、英国、德国和法国均在确诊病例数排名的前20位。
3.2 研究热点分析
3.2.1 主题聚类分析
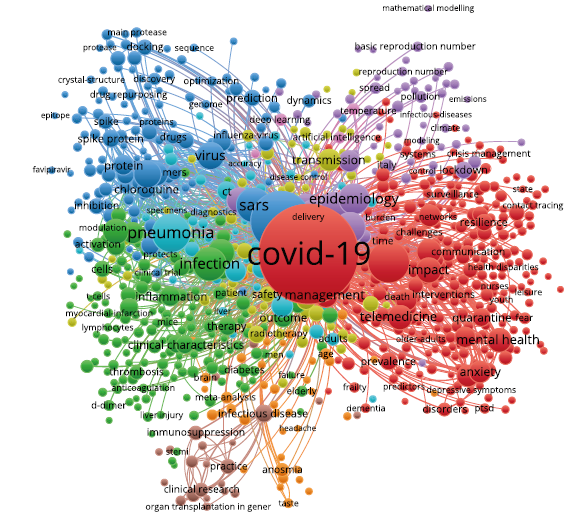
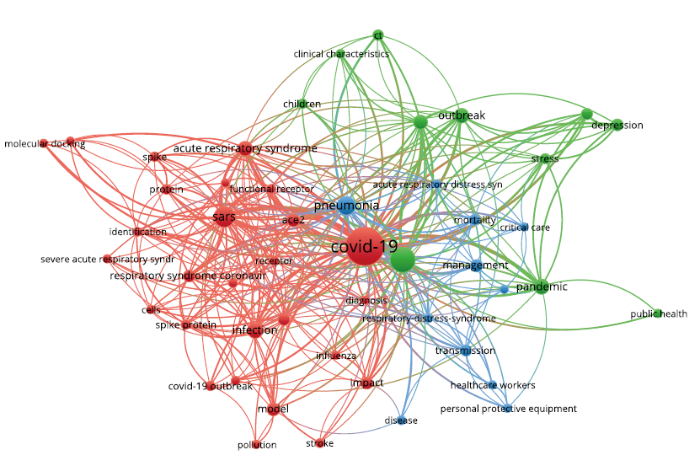
高频关键词的共现可以揭示出研究领域的热点主题,有利于分析某个领域的研究热点和前沿。利用VOSviewer进行关键词共现分析并聚类,分析类型选择Co-occurrence,分析单元选择all keywords。VOSviewer提供用于数据清理的词典操作功能,具体是通过复写官方提供的thesaurus_terms.txt文件进行清洗,可过滤掉同义词、单复数词汇、缩写词汇等表达形式不同的词汇以及词频高但意义不大的停用关键词等,共筛选出21 025个关键词。最终选取词频≥10的834个关键词进行分析,筛选出9个类别,绘制出相应的关键词共现聚类网络图(图4)。
图4
聚类图谱中,不同颜色代表不同的类别,标签的大小代表关键词的频率,标签越大表示关键词的频率越高。依据类别中频率较大的主题特征词,为每个类别命名相应的主题名称(表3),总结得到以下9个方面的研究热点。
表3 新冠肺炎研究的9个聚类主题
| 聚类号 | 聚类主题 | 子主题 | 颜色标识 | 主题特征词 | 代表性文献 (被引频次) |
|---|---|---|---|---|---|
| 1 | 新冠肺炎暴发及公共管理 | 疫情的暴发、流行病学、心理健康、社交媒体 | 红色 | COVID-19, pandemic, impact, management, mental health, anxiety, depression, public health, social media, policy | [3](1923)、[4](7679)、[5](257)、[7](735)、[8](623) |
| 2 | 临床特征与病理机制研究 | 临床特点、急性呼吸系统综合症、转化酶、受体、炎症、表达、细胞因子风暴 | 草绿 | clinical characteristics, acute respiratory syndrome, ACE2, receptor, inflammation, expression, cytokine storm | [9](4344)、[10](4807)、[19](1764) |
| 3 | 溯源、疫苗与治疗方法 | SARS病毒相关、疫苗、起源、氯喹、药物、抑制、分子对接 | 深蓝 | SARS, virus, vaccine, origin, chloroquine, drugs, inhibition, molecular docking | [20](3455)、[21](2296)、[22](1500)、[23](48) |
| 4 | 预防指南 | 传播、个人防护设备、预防、消毒、安全 | 黄色 | transmission, personal protective equipment, prevention, guidelines, healthcare workers | [30](3)、[31](11)、[32](13) |
| 5 | 流行病学的传播模型研究 | 模型、基本感染数R0、繁殖数 | 紫色 | SEIR model, sir model, mathematical model, spread, basic reproduction number, reproduction number | [26](2870)、[33](768) |
| 6 | 诊断方法研究 | 诊断、CT、症状诊断 | 天蓝 | pneumonia, diagnosis, CT, features | [34](855) |
| 7 | 交叉症状研究 | 嗅觉缺失、味觉障碍、耳鼻喉科、眼科 | 橙色 | anosmia, smell, olfaction , taste, dysgeusia, ent, ophthalmology | [37](48)、[38](102) |
| 8 | 临床实践及特殊群体的免疫抑制冲突 | 临床研究、实践、免疫抑制、肾脏移植 | 棕色 | clinical research, immunosuppression, kidney transplantation | [39](62) |
| 9 | 信息技术应用 | 人工智能、深度学习、机器学习 | 深绿 | deep learning, artificial intelligence, machine learning, classification | [40](17)、[41](30)、[44](120)、[45](36) |
(1)新冠肺炎的暴发及公共管理,涉及暴发初期、相关流行病学的研究,以及公共卫生、公众心理的管理;
(2)临床特征与病理机制研究,涉及临床症状、急性呼吸综合征研究及相关病理机制,如受体表达、细胞因子风暴的研究;
(3)溯源、疫苗研究及治疗方法,涉及和SARS病毒相关的溯源研究、疫苗进展及相关的治疗方法,如药物治疗、恢复期血浆治疗和疫苗方面的研究;
(4)新冠肺炎的预防指南,涉及预防新冠肺炎的传播和个人防护设备;
(5)流行病学传播模型的研究,涉及病毒传播方式的计算机模型;
(6)新冠肺炎的诊断方法研究,涉及CT诊断法、核酸检测诊断或依据临床特征的临床诊断方法;
(7)交叉症状研究,涉及耳鼻喉科、眼科相关交叉科室,如对患者嗅觉、味觉缺失的研究;
(8)新冠肺炎的临床实践研究,涉及临床实践、免疫抑制剂的研究,主要针对如肾脏移植患者等需要抑制免疫功能的特殊群体;
(9)信息技术在疫情期间的应用,如机器学习、深度学习等人工智能技术。
3.2.2 代表性文献
根据3.2.1小节聚类的主题特征词,选取相关主题下的代表性文献(表3),以辅助解读不同主题的阶段性研究进展。文献选取遵循以下原则:文献的关键词包含在聚类主题的主题特征词列表中;文献的被引频次相对较高(被引频次的统计截至2020-12-06)。
(1)新冠肺炎的暴发及公共管理
(2)临床特征与病理机制研究
(3)溯源、疫苗与治疗方法研究
在疫苗研发方面,新冠肺炎的疫苗研究有5个路线:全病毒灭活疫苗、基因工程亚单位疫苗(重组蛋白疫苗)、腺病毒载体疫苗、减毒流感病毒载体疫苗以及核酸疫苗。疫苗正式接种需要三期临床试验,1期临床是安全性指标的观察,2期临床是免疫原性和安全性指标的观察,3期临床主要在流行的人群、区域中观察疫苗是否能够预防感染,完成三期临床是疫苗研发成功的关键要素。2020年5月22日,由陈薇院士团队研制的腺病毒载体新冠疫苗已完成1期2期接种,1期结果的初步评价表明该疫苗接种28天后耐受性和免疫原性良好[27]。该疫苗成为我国首个获批进入临床研究的新冠疫苗,也是全球第一个进入2期临床试验的新冠疫苗,于2020年8月11日获得专利授权[28]。我国已于7月22日正式启动新冠疫苗的紧急使用,成为全球研发新冠病毒疫苗成效最为显著的国家。截至2020年8月17日世界卫生组织统计,全球共有170款候选疫苗,31款疫苗进入临床试验阶段,其中已有7款进入临床3期,4款来自中国,2款来自美国,1款来自英国[29],疫苗研发仍处于激烈的竞争阶段。且在我国的4款疫苗中,3款是灭活疫苗,1款是腺病毒载体疫苗,另外3种类型疫苗尚没有进入临床3期;英国1款疫苗是腺病毒载体疫苗;美国2款疫苗都属于核酸疫苗。因此,在疫苗研发领域,中国仍需并行推进不同技术路线的疫苗研究,以根据其不同优缺点,应对复杂的疫情需求。
(4)预防指南
(5)流行病学传播模型方面
(6)新冠肺炎的诊断方法研究
(7)交叉症状研究
(8)新冠肺炎的临床实践研究
牛津大学对新冠肺炎患者基于一种常见免疫抑制药物(类固醇地塞米松,Dexamethasone)的试验发现,对重病患者使用这种廉价类固醇药物可显著降低死亡率[39]。
(9)信息技术在疫情期间的应用
对新冠肺炎的传播进行预测:Yang等[44]据2003年SARS的数据进行训练构建修正了SEIR和AI预测的预测模型,可用于预测疫情发展;
实施数据监测、大规模数据处理:Allam等[45]建议使用大数据和人工智能技术构建智慧城市网络,未来在应对突发事件时可共享生物医疗数据,进行公共卫生监测;
机器人应用:机器人在医院、机场、交通系统、娱乐和景区、酒店等应用,有助于新冠肺炎期间的管理[46];
药物筛选:依据SARS-CoV-2与SARS-CoV高度同源特征,Zhang等[47]提出了一种基于深度学习的方法,可进行大规模的虚拟筛选和序列分析,研究结果提供了COVID-19 3C样蛋白酶的潜在药物清单。
3.2.3 中国与其他国家的合作主题分析
图5
表4 与中国的合作强度Top10国家
| 序号 | 国家 | 链接强度 |
|---|---|---|
| 1 | 美国 | 448 |
| 2 | 英国 | 178 |
| 3 | 澳大利亚 | 103 |
| 4 | 加拿大 | 95 |
| 5 | 意大利 | 87 |
| 6 | 印度 | 82 |
| 7 | 法国 | 51 |
| 8 | 泰国 | 49 |
| 9 | 德国 | 49 |
| 10 | 新加坡 | 44 |
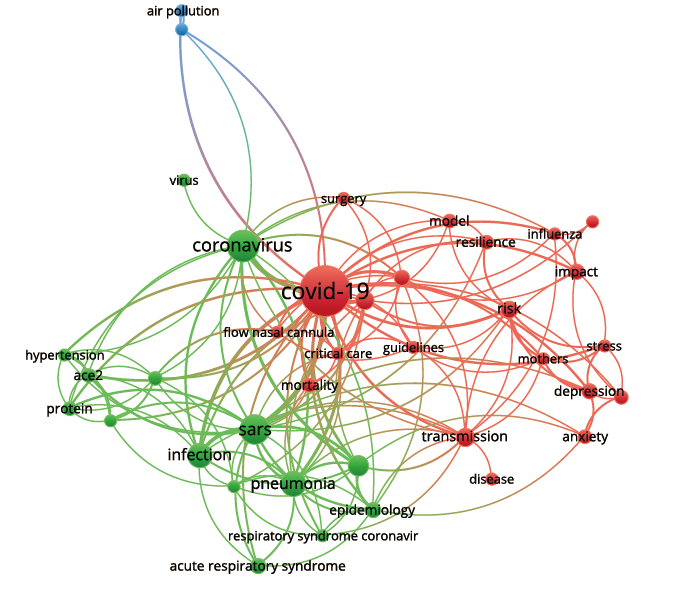
(1)中-美文献的聚类主题
图6
表5 中-美合作论文的3个主题聚类
| 聚类号 | 聚类主题 | 子主题 | 颜色标识 | 主题特征词 |
|---|---|---|---|---|
| 1 | 病理机制与感染研究 | SARS、转化酶、急性呼吸综合征、感染、模型 | 红色 | COVID-19、SARS、ACE2、acute respiratory syndrome、infection、protein、function recepter |
| 2 | 心理健康研究 | 公共健康、压力 | 绿色 | pneumonia 、public health、stress、depression |
| 3 | 传播与防护管理 | 传播、管理医护人员、个人保护设备 | 蓝色 | transmission、management、healthcare workers、personal protective equipment |
聚类结果图显示中美合作的研究主题相对较分散,故将关联紧密的主题合并,归纳为3部分,主要包括:(i)新冠疫情的病理机制研究,(ii)公共心理健康研究,(iii)传播机制与防护管理。图中这3个部分之间连线较多、距离较近,说明主题之间的关系紧密。
(2)中-英文献的聚类主题
图7
表6 中-英合作论文的3个主题聚类
| 聚类号 | 聚类主题 | 子主题 | 颜色标识 | 主题特征词 |
|---|---|---|---|---|
| 1 | 病理机制研究 | SARS、急性呼吸系统症状、流行病学 | 绿色 | coronavirus、SARS、pneumonia、acute respiratory syndrome、epidemiology、ACE2、expression |
| 2 | 疫情防控管理 | 感染、传播、管理、影响 | 红色 | COVID-19、pandemic、infection、pneumonia、outbreak、wuhan、transmission、guidlines |
| 3 | 空气污染的影响 | 空气污染、PM2.5 | 蓝色 | PM2.5、air pollution |
聚类结果图中可以发现中英合作的研究主题同样归为3类,但子主题相对中美合作较少。研究主题主要包括:(i)病理机制研究,(ii)疫情防控管理,(iii)空气污染与COVID-19之间的相互影响。
4 结论
本文使用文献计量分析方法,利用VOSviewer绘制全球关于新冠肺炎的研究热点主题图谱和合作图谱,可视化展示研究热点和国际合作的情况。初步归纳出全球关于新冠肺炎的研究热点主题和需要加强合作的方向,可为研究学者和政策制定者提供参考建议,主要结论如下。
第一,在2019年12月1日至2020年8月25日期间,关于新冠肺炎的研究热点集中于预防与公共卫生及公众心理管理,临床特征与病理机制研究、疫苗和治疗方法研究、预防指南、传播模型、诊断方法、交叉症状、临床实践、信息技术的辅助应用这几个方面。中国研究者在本阶段关于新冠肺炎的研究发表了较多代表性的成果。
第二,中国的发文量仅次于美国,排名第2,独立完成的论文占比位居全球首位,但另一方面,国际合作的总连接强度位于全球第4,低于美国、英国和意大利。因此,中国在下一阶段新冠肺炎研究方面,可以考虑加强与其他国家的合作。同时,面对当前日渐紧张的世界战略竞争环境,尤其是美国对中国的遏制和技术封锁,中国需要专注于独创性的技术攻关和科技创新。
第三,对比全球的新冠肺炎研究主题与中-美、中-英的合作研究主题发现,中国和美、英等国可在治疗方法、临床实践、交叉症状、模型预测等主题的研究上开展更多的科研交流,进一步加强合作。
第四,新冠肺炎领域还有很多研究尚无准确的定论,如新冠肺炎的中间宿主问题、核酸检测的假阴性问题,以及丧失嗅觉或味觉的症状是否可以纳入新冠肺炎的症状中等,围绕此类问题已开展很多研究,但未有准确的定论,有待科研工作者深入探索。
通过本研究的文献计量分析发现,我国在新冠肺炎相关研究方面取得了多项研究成果,在科技抗疫方面发挥了关键支撑作用。结合我国在新冠疫苗、药物研发和临床救治等诸多方面取得的实践性突破,尤其是在灭活类型疫苗上的领跑,体现了我国的科研创新能力和大国表率作用。另外,本研究文集的时间范围为2019–2020年,时间跨度不足9个月,不适合进行更细粒度的时间演变分析,随着后续研究的不断深入,有待开展更为细化和全面的研究。
参考文献
Software survey: VOSviewer, a computer program for bibliometric mapping
[J].We present VOSviewer, a freely available computer program that we have developed for constructing and viewing bibliometric maps. Unlike most computer programs that are used for bibliometric mapping, VOSviewer pays special attention to the graphical representation of bibliometric maps. The functionality of VOSviewer is especially useful for displaying large bibliometric maps in an easy-to-interpret way. The paper consists of three parts. In the first part, an overview of VOSviewer's functionality for displaying bibliometric maps is provided. In the second part, the technical implementation of specific parts of the program is discussed. Finally, in the third part, VOSviewer's ability to handle large maps is demonstrated by using the program to construct and display a co-citation map of 5,000 major scientific journals.
A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster
[J].BACKGROUND: An ongoing outbreak of pneumonia associated with a novel coronavirus was reported in Wuhan city, Hubei province, China. Affected patients were geographically linked with a local wet market as a potential source. No data on person-to-person or nosocomial transmission have been published to date. METHODS: In this study, we report the epidemiological, clinical, laboratory, radiological, and microbiological findings of five patients in a family cluster who presented with unexplained pneumonia after returning to Shenzhen, Guangdong province, China, after a visit to Wuhan, and an additional family member who did not travel to Wuhan. Phylogenetic analysis of genetic sequences from these patients were done. FINDINGS: From Jan 10, 2020, we enrolled a family of six patients who travelled to Wuhan from Shenzhen between Dec 29, 2019 and Jan 4, 2020. Of six family members who travelled to Wuhan, five were identified as infected with the novel coronavirus. Additionally, one family member, who did not travel to Wuhan, became infected with the virus after several days of contact with four of the family members. None of the family members had contacts with Wuhan markets or animals, although two had visited a Wuhan hospital. Five family members (aged 36-66 years) presented with fever, upper or lower respiratory tract symptoms, or diarrhoea, or a combination of these 3-6 days after exposure. They presented to our hospital (The University of Hong Kong-Shenzhen Hospital, Shenzhen) 6-10 days after symptom onset. They and one asymptomatic child (aged 10 years) had radiological ground-glass lung opacities. Older patients (aged >60 years) had more systemic symptoms, extensive radiological ground-glass lung changes, lymphopenia, thrombocytopenia, and increased C-reactive protein and lactate dehydrogenase levels. The nasopharyngeal or throat swabs of these six patients were negative for known respiratory microbes by point-of-care multiplex RT-PCR, but five patients (four adults and the child) were RT-PCR positive for genes encoding the internal RNA-dependent RNA polymerase and surface Spike protein of this novel coronavirus, which were confirmed by Sanger sequencing. Phylogenetic analysis of these five patients' RT-PCR amplicons and two full genomes by next-generation sequencing showed that this is a novel coronavirus, which is closest to the bat severe acute respiatory syndrome (SARS)-related coronaviruses found in Chinese horseshoe bats. INTERPRETATION: Our findings are consistent with person-to-person transmission of this novel coronavirus in hospital and family settings, and the reports of infected travellers in other geographical regions. FUNDING: The Shaw Foundation Hong Kong, Michael Seak-Kan Tong, Respiratory Viral Research Foundation Limited, Hui Ming, Hui Hoy and Chow Sin Lan Charity Fund Limited, Marina Man-Wai Lee, the Hong Kong Hainan Commercial Association South China Microbiology Research Fund, Sanming Project of Medicine (Shenzhen), and High Level-Hospital Program (Guangdong Health Commission).
Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China
[J].BACKGROUND: A recent cluster of pneumonia cases in Wuhan, China, was caused by a novel betacoronavirus, the 2019 novel coronavirus (2019-nCoV). We report the epidemiological, clinical, laboratory, and radiological characteristics and treatment and clinical outcomes of these patients. METHODS: All patients with suspected 2019-nCoV were admitted to a designated hospital in Wuhan. We prospectively collected and analysed data on patients with laboratory-confirmed 2019-nCoV infection by real-time RT-PCR and next-generation sequencing. Data were obtained with standardised data collection forms shared by WHO and the International Severe Acute Respiratory and Emerging Infection Consortium from electronic medical records. Researchers also directly communicated with patients or their families to ascertain epidemiological and symptom data. Outcomes were also compared between patients who had been admitted to the intensive care unit (ICU) and those who had not. FINDINGS: By Jan 2, 2020, 41 admitted hospital patients had been identified as having laboratory-confirmed 2019-nCoV infection. Most of the infected patients were men (30 [73%] of 41); less than half had underlying diseases (13 [32%]), including diabetes (eight [20%]), hypertension (six [15%]), and cardiovascular disease (six [15%]). Median age was 49.0 years (IQR 41.0-58.0). 27 (66%) of 41 patients had been exposed to Huanan seafood market. One family cluster was found. Common symptoms at onset of illness were fever (40 [98%] of 41 patients), cough (31 [76%]), and myalgia or fatigue (18 [44%]); less common symptoms were sputum production (11 [28%] of 39), headache (three [8%] of 38), haemoptysis (two [5%] of 39), and diarrhoea (one [3%] of 38). Dyspnoea developed in 22 (55%) of 40 patients (median time from illness onset to dyspnoea 8.0 days [IQR 5.0-13.0]). 26 (63%) of 41 patients had lymphopenia. All 41 patients had pneumonia with abnormal findings on chest CT. Complications included acute respiratory distress syndrome (12 [29%]), RNAaemia (six [15%]), acute cardiac injury (five [12%]) and secondary infection (four [10%]). 13 (32%) patients were admitted to an ICU and six (15%) died. Compared with non-ICU patients, ICU patients had higher plasma levels of IL2, IL7, IL10, GSCF, IP10, MCP1, MIP1A, and TNFalpha. INTERPRETATION: The 2019-nCoV infection caused clusters of severe respiratory illness similar to severe acute respiratory syndrome coronavirus and was associated with ICU admission and high mortality. Major gaps in our knowledge of the origin, epidemiology, duration of human transmission, and clinical spectrum of disease need fulfilment by future studies. FUNDING: Ministry of Science and Technology, Chinese Academy of Medical Sciences, National Natural Science Foundation of China, and Beijing Municipal Science and Technology Commission.
Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20-28 January 2020
[J].
Clinical characteristics of 2019 novel coronavirus infection in China
[J].
Immediate psychological responses and associated factors during the initial stage of the 2019 coronavirus disease (COVID-19) epidemic among the general population in China
[J].
Factors associated with mental health outcomes among health care workers exposed to coronavirus disease 2019
[J].
Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study
[J].BACKGROUND: In December, 2019, a pneumonia associated with the 2019 novel coronavirus (2019-nCoV) emerged in Wuhan, China. We aimed to further clarify the epidemiological and clinical characteristics of 2019-nCoV pneumonia. METHODS: In this retrospective, single-centre study, we included all confirmed cases of 2019-nCoV in Wuhan Jinyintan Hospital from Jan 1 to Jan 20, 2020. Cases were confirmed by real-time RT-PCR and were analysed for epidemiological, demographic, clinical, and radiological features and laboratory data. Outcomes were followed up until Jan 25, 2020. FINDINGS: Of the 99 patients with 2019-nCoV pneumonia, 49 (49%) had a history of exposure to the Huanan seafood market. The average age of the patients was 55.5 years (SD 13.1), including 67 men and 32 women. 2019-nCoV was detected in all patients by real-time RT-PCR. 50 (51%) patients had chronic diseases. Patients had clinical manifestations of fever (82 [83%] patients), cough (81 [82%] patients), shortness of breath (31 [31%] patients), muscle ache (11 [11%] patients), confusion (nine [9%] patients), headache (eight [8%] patients), sore throat (five [5%] patients), rhinorrhoea (four [4%] patients), chest pain (two [2%] patients), diarrhoea (two [2%] patients), and nausea and vomiting (one [1%] patient). According to imaging examination, 74 (75%) patients showed bilateral pneumonia, 14 (14%) patients showed multiple mottling and ground-glass opacity, and one (1%) patient had pneumothorax. 17 (17%) patients developed acute respiratory distress syndrome and, among them, 11 (11%) patients worsened in a short period of time and died of multiple organ failure. INTERPRETATION: The 2019-nCoV infection was of clustering onset, is more likely to affect older males with comorbidities, and can result in severe and even fatal respiratory diseases such as acute respiratory distress syndrome. In general, characteristics of patients who died were in line with the MuLBSTA score, an early warning model for predicting mortality in viral pneumonia. Further investigation is needed to explore the applicability of the MuLBSTA score in predicting the risk of mortality in 2019-nCoV infection. FUNDING: National Key R&D Program of China.
Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-infected pneumonia in Wuhan, China
[J].
Clinical characteristics and outcome of SARS-CoV-2-infected patients with haematological diseases: a retrospective case study in four hospitals in Italy, Spain and the Netherlands
[J].
Clinical Characteristics of Covid-19 in New York city
[J].
Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China
[J].
Epidemiologic and Clinical Characteristics of Novel Coronavirus Infections Involving 13 patients outside Wuhan, China
[J].
SARS-CoV-2 infection in children: transmission dynamics and clinical characteristics
[J].
Clinical characteristics and outcomes of COVID-19 in solid organ transplant recipients: A case-control study
[J].Solid organ transplant recipients (SOTr) with coronavirus disease 2019 (COVID-19) are expected to have poorer outcomes compared to nontransplant patients because of immunosuppression and comorbidities. The clinical characteristics of 47 SOTr (38 kidneys and 9 nonkidney organs) were compared to 100 consecutive hospitalized nontransplant controls. Twelve of 47 SOTr managed as outpatients were subsequently excluded from the outcome analyses to avoid potential selection bias. Chronic kidney disease (89% vs 57% P = .0007), diabetes (66% vs 33% P = .0007), and hypertension (94% vs 72% P = .006) were more common in the 35 hospitalized SOTr compared to controls. Diarrhea (54% vs 17%, P < .0001) was more frequent in SOTr. Primary composite outcome (escalation to intensive care unit, mechanical ventilation, or in-hospital all-cause mortality) was comparable between SOTr and controls (40% vs 48%, odds ratio [OR] 0.72 confidence interval [CI] [0.33-1.58] P = .42), despite more comorbidities in SOTr. Acute kidney injury requiring renal replacement therapy occurred in 20% of SOTr compared to 4% of controls (OR 6 CI [1.64-22] P = .007). Multivariate analysis demonstrated that increasing age and clinical severity were associated with mortality. Transplant status itself was not associated with mortality.
Clinical characteristics of pregnant women with Covid-19 in Wuhan, China
[J].
COVID-19 and pregnancy: A review of clinical characteristics, obstetric outcomes and vertical transmission
[J].BACKGROUND: Since its emergence in December 2019, COVID-19 has spread to over 210 countries, with an estimated mortality rate of 3-4%. Little is understood about its effects during pregnancy. AIMS: To describe the current understanding of COVID-19 illness in pregnant women, to describe obstetric outcomes and to identify gaps in the existing knowledge. METHODS: Medline Ovid, EMBASE, World Health Organization COVID-19 research database and Cochrane COVID-19 in pregnancy spreadsheet were accessed on 18/4, 18/5 and 23/5 2020. Articles were screened via Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines. The following were excluded: reviews, opinion pieces, guidelines, articles pertaining solely to other viruses, single case reports. RESULTS: Sixty articles were included in this review. Some pregnant participants may have been included in multiple publications, as admission dates overlap for reports from the same hospital. However, a total of 1287 confirmed SARS-CoV-2 positive pregnant cases are reported. Where universal testing was undertaken, asymptomatic infection occurred in 43.5-92% of cases. In the cohort studies, severe and critical COVID-19 illness rates approximated those of the non-pregnant population. Eight maternal deaths, six neonatal deaths, seven stillbirths and five miscarriages were reported. Nineteen neonates were SARS-CoV-2 positive, confirmed by reverse transcription polymerase chain reaction of nasopharyngeal swabs. [Correction added on 2 September 2020, after first online publication: the number of neonates indicated in the preceding sentence has been corrected from 'Thirteen' to 'Nineteen'.] CONCLUSIONS: Where universal screening was conducted, SARS-CoV-2 infection in pregnancy was often asymptomatic. Severe and critical disease rates approximate those in the general population. Vertical transmission is possible; however, it is unclear whether SARS-CoV-2 positive neonates were infected in utero, intrapartum or postpartum. Future work should assess risks of congenital syndromes and adverse perinatal outcomes where infection occurs in early and mid-pregnancy.
Pathological findings of COVID-19 associated with acute respiratory distress syndrome
[J].
A pneumonia outbreak associated with a new coronavirus of probable bat origin
[J].Since the outbreak of severe acute respiratory syndrome (SARS) 18 years ago, a large number of SARS-related coronaviruses (SARSr-CoVs) have been discovered in their natural reservoir host, bats(1-4). Previous studies have shown that some bat SARSr-CoVs have the potential to infect humans(5-7). Here we report the identification and characterization of a new coronavirus (2019-nCoV), which caused an epidemic of acute respiratory syndrome in humans in Wuhan, China. The epidemic, which started on 12 December 2019, had caused 2,794 laboratory-confirmed infections including 80 deaths by 26 January 2020. Full-length genome sequences were obtained from five patients at an early stage of the outbreak. The sequences are almost identical and share 79.6% sequence identity to SARS-CoV. Furthermore, we show that 2019-nCoV is 96% identical at the whole-genome level to a bat coronavirus. Pairwise protein sequence analysis of seven conserved non-structural proteins domains show that this virus belongs to the species of SARSr-CoV. In addition, 2019-nCoV virus isolated from the bronchoalveolar lavage fluid of a critically ill patient could be neutralized by sera from several patients. Notably, we confirmed that 2019-nCoV uses the same cell entry receptor-angiotensin converting enzyme II (ACE2)-as SARS-CoV.
Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding
[J].BACKGROUND: In late December, 2019, patients presenting with viral pneumonia due to an unidentified microbial agent were reported in Wuhan, China. A novel coronavirus was subsequently identified as the causative pathogen, provisionally named 2019 novel coronavirus (2019-nCoV). As of Jan 26, 2020, more than 2000 cases of 2019-nCoV infection have been confirmed, most of which involved people living in or visiting Wuhan, and human-to-human transmission has been confirmed. METHODS: We did next-generation sequencing of samples from bronchoalveolar lavage fluid and cultured isolates from nine inpatients, eight of whom had visited the Huanan seafood market in Wuhan. Complete and partial 2019-nCoV genome sequences were obtained from these individuals. Viral contigs were connected using Sanger sequencing to obtain the full-length genomes, with the terminal regions determined by rapid amplification of cDNA ends. Phylogenetic analysis of these 2019-nCoV genomes and those of other coronaviruses was used to determine the evolutionary history of the virus and help infer its likely origin. Homology modelling was done to explore the likely receptor-binding properties of the virus. FINDINGS: The ten genome sequences of 2019-nCoV obtained from the nine patients were extremely similar, exhibiting more than 99.98% sequence identity. Notably, 2019-nCoV was closely related (with 88% identity) to two bat-derived severe acute respiratory syndrome (SARS)-like coronaviruses, bat-SL-CoVZC45 and bat-SL-CoVZXC21, collected in 2018 in Zhoushan, eastern China, but were more distant from SARS-CoV (about 79%) and MERS-CoV (about 50%). Phylogenetic analysis revealed that 2019-nCoV fell within the subgenus Sarbecovirus of the genus Betacoronavirus, with a relatively long branch length to its closest relatives bat-SL-CoVZC45 and bat-SL-CoVZXC21, and was genetically distinct from SARS-CoV. Notably, homology modelling revealed that 2019-nCoV had a similar receptor-binding domain structure to that of SARS-CoV, despite amino acid variation at some key residues. INTERPRETATION: 2019-nCoV is sufficiently divergent from SARS-CoV to be considered a new human-infecting betacoronavirus. Although our phylogenetic analysis suggests that bats might be the original host of this virus, an animal sold at the seafood market in Wuhan might represent an intermediate host facilitating the emergence of the virus in humans. Importantly, structural analysis suggests that 2019-nCoV might be able to bind to the angiotensin-converting enzyme 2 receptor in humans. The future evolution, adaptation, and spread of this virus warrant urgent investigation. FUNDING: National Key Research and Development Program of China, National Major Project for Control and Prevention of Infectious Disease in China, Chinese Academy of Sciences, Shandong First Medical University.
A new coronavirus associated with human respiratory disease in China
[J].Emerging infectious diseases, such as severe acute respiratory syndrome (SARS) and Zika virus disease, present a major threat to public health(1-3). Despite intense research efforts, how, when and where new diseases appear are still a source of considerable uncertainty. A severe respiratory disease was recently reported in Wuhan, Hubei province, China. As of 25 January 2020, at least 1,975 cases had been reported since the first patient was hospitalized on 12 December 2019. Epidemiological investigations have suggested that the outbreak was associated with a seafood market in Wuhan. Here we study a single patient who was a worker at the market and who was admitted to the Central Hospital of Wuhan on 26 December 2019 while experiencing a severe respiratory syndrome that included fever, dizziness and a cough. Metagenomic RNA sequencing(4) of a sample of bronchoalveolar lavage fluid from the patient identified a new RNA virus strain from the family Coronaviridae, which is designated here 'WH-Human 1' coronavirus (and has also been referred to as '2019-nCoV'). Phylogenetic analysis of the complete viral genome (29,903 nucleotides) revealed that the virus was most closely related (89.1% nucleotide similarity) to a group of SARS-like coronaviruses (genus Betacoronavirus, subgenus Sarbecovirus) that had previously been found in bats in China(5). This outbreak highlights the ongoing ability of viral spill-over from animals to cause severe disease in humans.
Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins
[J].
Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro
[J].
Treatment of 5 Critically 3 Patients with COVID-19 with convalescent plasma
[J].
Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia
[J].
Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial
[J].
Preventing transmission among operating room staff during COVID-19 pandemic: the role of the Aerosol Box and other personal protective equipment
[J].
Opinion to address the personal protective equipment shortage in the global community during the COVID-19 outbreak
[J].
Electronic personal protective equipment: A strategy to protect emergency department providers in the age of COVID-19
[J].
Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study
[J].BACKGROUND: Since Dec 31, 2019, the Chinese city of Wuhan has reported an outbreak of atypical pneumonia caused by the 2019 novel coronavirus (2019-nCoV). Cases have been exported to other Chinese cities, as well as internationally, threatening to trigger a global outbreak. Here, we provide an estimate of the size of the epidemic in Wuhan on the basis of the number of cases exported from Wuhan to cities outside mainland China and forecast the extent of the domestic and global public health risks of epidemics, accounting for social and non-pharmaceutical prevention interventions. METHODS: We used data from Dec 31, 2019, to Jan 28, 2020, on the number of cases exported from Wuhan internationally (known days of symptom onset from Dec 25, 2019, to Jan 19, 2020) to infer the number of infections in Wuhan from Dec 1, 2019, to Jan 25, 2020. Cases exported domestically were then estimated. We forecasted the national and global spread of 2019-nCoV, accounting for the effect of the metropolitan-wide quarantine of Wuhan and surrounding cities, which began Jan 23-24, 2020. We used data on monthly flight bookings from the Official Aviation Guide and data on human mobility across more than 300 prefecture-level cities in mainland China from the Tencent database. Data on confirmed cases were obtained from the reports published by the Chinese Center for Disease Control and Prevention. Serial interval estimates were based on previous studies of severe acute respiratory syndrome coronavirus (SARS-CoV). A susceptible-exposed-infectious-recovered metapopulation model was used to simulate the epidemics across all major cities in China. The basic reproductive number was estimated using Markov Chain Monte Carlo methods and presented using the resulting posterior mean and 95% credibile interval (CrI). FINDINGS: In our baseline scenario, we estimated that the basic reproductive number for 2019-nCoV was 2.68 (95% CrI 2.47-2.86) and that 75 815 individuals (95% CrI 37 304-130 330) have been infected in Wuhan as of Jan 25, 2020. The epidemic doubling time was 6.4 days (95% CrI 5.8-7.1). We estimated that in the baseline scenario, Chongqing, Beijing, Shanghai, Guangzhou, and Shenzhen had imported 461 (95% CrI 227-805), 113 (57-193), 98 (49-168), 111 (56-191), and 80 (40-139) infections from Wuhan, respectively. If the transmissibility of 2019-nCoV were similar everywhere domestically and over time, we inferred that epidemics are already growing exponentially in multiple major cities of China with a lag time behind the Wuhan outbreak of about 1-2 weeks. INTERPRETATION: Given that 2019-nCoV is no longer contained within Wuhan, other major Chinese cities are probably sustaining localised outbreaks. Large cities overseas with close transport links to China could also become outbreak epicentres, unless substantial public health interventions at both the population and personal levels are implemented immediately. Independent self-sustaining outbreaks in major cities globally could become inevitable because of substantial exportation of presymptomatic cases and in the absence of large-scale public health interventions. Preparedness plans and mitigation interventions should be readied for quick deployment globally. FUNDING: Health and Medical Research Fund (Hong Kong, China).
Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR
[J].
Smell and taste dysfunction in patients with COVID-19
[J].
Alterations in smell or taste in mildly symptomatic outpatients with SARS-CoV-2 infection
[J].
Coronavirus breakthrough: dexamethasone is first drug shown to save lives
[J].
Deep learning system to screen coronavirus disease 2019 pneumonia
[J].
Artificial intelligence-enabled rapid diagnosis of patients with COVID-19
[J].For diagnosis of coronavirus disease 2019 (COVID-19), a SARS-CoV-2 virus-specific reverse transcriptase polymerase chain reaction (RT-PCR) test is routinely used. However, this test can take up to 2 d to complete, serial testing may be required to rule out the possibility of false negative results and there is currently a shortage of RT-PCR test kits, underscoring the urgent need for alternative methods for rapid and accurate diagnosis of patients with COVID-19. Chest computed tomography (CT) is a valuable component in the evaluation of patients with suspected SARS-CoV-2 infection. Nevertheless, CT alone may have limited negative predictive value for ruling out SARS-CoV-2 infection, as some patients may have normal radiological findings at early stages of the disease. In this study, we used artificial intelligence (AI) algorithms to integrate chest CT findings with clinical symptoms, exposure history and laboratory testing to rapidly diagnose patients who are positive for COVID-19. Among a total of 905 patients tested by real-time RT-PCR assay and next-generation sequencing RT-PCR, 419 (46.3%) tested positive for SARS-CoV-2. In a test set of 279 patients, the AI system achieved an area under the curve of 0.92 and had equal sensitivity as compared to a senior thoracic radiologist. The AI system also improved the detection of patients who were positive for COVID-19 via RT-PCR who presented with normal CT scans, correctly identifying 17 of 25 (68%) patients, whereas radiologists classified all of these patients as COVID-19 negative. When CT scans and associated clinical history are available, the proposed AI system can help to rapidly diagnose COVID-19 patients.
Deep learning for detecting corona virus disease 2019 (COVID-19) on high-resolution computed tomography: a pilot study
[J].
Towards an artificial intelligence framework for data-driven prediction of coronavirus clinical severity
[J].
Modified SEIR and AI prediction of the epidemics trend of COVID-19 in China under public health interventions
[J].Background: The coronavirus disease 2019 (COVID-19) outbreak originating in Wuhan, Hubei province, China, coincided with chunyun, the period of mass migration for the annual Spring Festival. To contain its spread, China adopted unprecedented nationwide interventions on January 23 2020. These policies included large-scale quarantine, strict controls on travel and extensive monitoring of suspected cases. However, it is unknown whether these policies have had an impact on the epidemic. We sought to show how these control measures impacted the containment of the epidemic. Methods: We integrated population migration data before and after January 23 and most updated COVID-19 epidemiological data into the Susceptible-Exposed-Infectious-Removed (SEIR) model to derive the epidemic curve. We also used an artificial intelligence (AI) approach, trained on the 2003 SARS data, to predict the epidemic. Results: We found that the epidemic of China should peak by late February, showing gradual decline by end of April. A five-day delay in implementation would have increased epidemic size in mainland China three-fold. Lifting the Hubei quarantine would lead to a second epidemic peak in Hubei province in mid-March and extend the epidemic to late April, a result corroborated by the machine learning prediction. Conclusions: Our dynamic SEIR model was effective in predicting the COVID-19 epidemic peaks and sizes. The implementation of control measures on January 23 2020 was indispensable in reducing the eventual COVID-19 epidemic size.
On the coronavirus (COVID-19) outbreak and the smart city nnetwork: vniversal data sharing standards coupled with artificial intelligence (AI) to benefit vrban health monitoring and management
[J].
From high-touch to high-tech: COVID-19 drives robotics adoption
[J].