植物自交不亲和性广泛存在于显花植物中,是防止近亲繁殖和促进开花植物遗传多样性和杂种优势的重要遗传机制。关于自交不亲和机制的研究多集中在S基因上。该文结合文献计量的方法,利用Hiscite、Citespace和VosViewer分析了基于S位点的植物自交不亲和机制研究的年代趋势、重点国家、重要机构、重要作者、研究前沿和研究热点,对蛋白质磷酸化、S-RNase泛素化和拟南芥属的演化作了简要论述,并根据目前的研究现状提出了该领域研究中存在的问题及解决对策,旨在为进一步研究提供重要参考依据。
Abstract:
Plant self-incompatibility is widely found in flowering plants. It’s an important genetic mechanism to prevent inbreeding and increase the genetic diversity and heterosis of flowering plant. About self-incompatibility mechanism, most of the researches focus on the S gene. In this paper, we use Hiscite, Citespace and VosViewer based on Bibliometrics to analyze self-incompatibility mechanism in the S-locus of plant, and have a brief discussion on protein phosphorylation, S-RNase ubiquitination and the evolution of Arabidopsis. According to the current research status in this field, we also put forward the existing problems and solutions to provide a reference for further study.
自交不亲和性(self-incompatibility, SI)是指具有完全花并能形成正常雌、雄配子,然而缺乏自花授粉结实能力的一种自交不育性,常存在于雌雄同株的植物,自交不亲和现象广泛分布在显花植物中[1]。SI是防止近亲繁殖和促进开花植物遗传多样性和杂种优势的重要遗传机制[2]。基于花粉识别特异性的遗传决定方式,SI可分为配子体型自交不亲和性(gametophytic self-incompatibility, GSI)和孢子体型自交不亲和性(sporophytic self-incompatibility, SSI) [3]。GSI即是花粉在柱头上发芽后可侵入柱头,且能在花柱组织中延伸一定的距离,之后便受到抑制。花粉管与雌性因素的抑制关系发生在单倍体配子体之间,常见于罂粟科(Papaveraceae)、蔷薇科(Rosaceae)、茄科(Solanaceae)和玄参科(Scrophulariaceae)的一些植物[4]。此种抑制关系可以发生在花柱组织中,也能发生在花粉管与胚囊组织之间,有的甚至是精子已经到达胚囊内,仍然不能与卵细胞结合[5]。SSI即是花粉落到柱头上不能正常发芽,或者发芽之后在柱头乳突细胞上缠绕而无法侵入柱头。这种不亲和关系发生在花粉管与柱头乳突细胞的孢子体之间,花粉的行为决定于二倍体亲本的基因型,多见于油菜、拟南芥等十字花科(Brassicaceae)植物中[6]。
植物受粉过程包括花粉落到柱头上、粘附、水合、萌发,花粉管生长、花粉到达胚珠完成受精作用几个阶段,植物自交不亲和反应的识别可以发生在受粉过程的任何一个阶段[7]。如拟南芥的识别发生在花粉水合阶段[8, 9];罂粟的不亲和反应发生在花粉萌发阶段[10];金鱼草的识别和不亲和反应都发生在花粉管内部[11]。不论是GSI还是SSI遗传上都是由特定的复等位基因控制,大多数植物的自交不亲和性是由单一的复等位基因位点控制,称为S位点(S-locus),S位点具有高度多态性[3]。少数植物的自交不亲和性是由两个及以上位点控制的,如禾本科的自交不亲和性由S和Z两个互不连锁的遗传位点共同控制[12]。S位点通常包含两类基因:决定花粉识别特异性的花粉S基因与决定花柱识别特异性的花柱S基因。蔷薇科、玄参科和茄科的花柱和花粉决定因子被鉴定为S-核酸酶(S-RNase)和F-box蛋白[13]。十字花科的花柱和花粉决定因子为S位点受体激酶 (S-locus receptor kinase , SRK)和SCR(S-locus cysteine-rich)[9]。而罂粟科的花柱和花粉决定因子为PrsS(Palaver rhoeas stigma S-determinant)和PrpS(Papaver rhoeas pollen S-determinant),且通过花粉管胞质自由Ca2+介导的信号级联激活作用来调控[14]。SI是研究有性植物生殖细胞间信号识别、转导以及细胞间相互作用的一种理想模型,在植物遗传改良和作物杂种优势利用上具有重要价值。
本文文献信息源自于汤森路透的Web of Science数据库核心合集,通过检索式TS= Self-Incompatibilit* and (s-rnase or f-box or S-locus or "s gene"),选取了1990–2016年的全部文献记录共计1 310条,数据库更新时间为2016年3月19日,其中2016年文献记录为4条。利用Hiscite、Citespace、Excel、VosViewer软件分析了有关植物S位点自交不亲和机制研究的年代趋势、重点国家、重要机构、重要作者、研究前沿与研究热点,旨在为进一步研究提供参考。
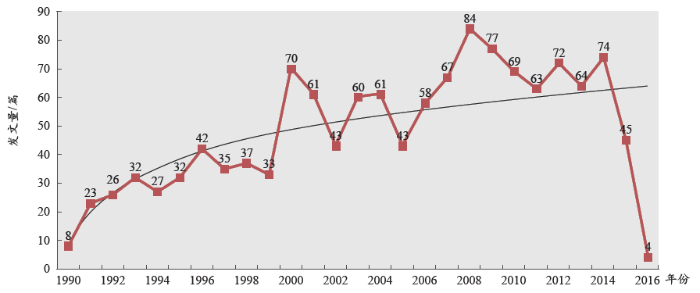
植物S位点自交不亲和机制年度发文量如图1所示。总体而言,植物自交不亲和机制研究领域发文量较少,自1990年开始呈现稳步上升的趋势,体现出该研究领域的重要意义和广泛的应用前景。2000年和2008年发文量呈现出两个波峰,其中2000年发文量的跃升是由于美国和日本发文量较前几年大幅度增加,两国发文量总和达到46篇之多,而2000年之前,美国和日本的年发文量仅为10篇左右。2015年和2016年发文量偏少,是由于数据库还未及时收录完整所致。
结合Histcite软件和Web of Science数据库对文献分布情况进行统计,在这26年内,Web of Science数据库中记录的发文数量前10位的国家如表1所示。在此期间,美国参与发表的论文数量为342篇,占发文总数的26.11%。其次是日本,占发文总数的24.58%,这两个国家是植物S位点自交不亲和机制研究的主力。中国和英格兰发文量排名紧随其后,但发文量均不到美国和日本的一半。从发表文章的被引用情况来看,美国的论文总被引用次数为15 273次,远远超过其他国家;日本次之,被引用次数为9 483次。二者的篇均被引用次数也显著高于其他国家。在检索出的1 310条记录中,被引次数≥50的论文总计220篇。中国的发文量虽然较多,但被引次数≥50的论文仅有9篇,与发达国家相比仍然存在较大差距。综合相对优势指数1(1 相对优势指数:假定国际篇均被引频次的平均水平为1,利用各国的篇均被引频次与其相对比,可以体现出各国的科研实力在国际上的相对水平。)和被引次数≥50的论文数量来看,美国、日本、英格兰、加拿大在自交不亲和机制研究领域中具有较大的学术影响力。
随着世界科技进入大科学时代,国家间的合作尤为重要。利用Citespace软件,采用Pathfinder算法对原始数据进行可视化,反映出植物S位点自交不亲和机制研究主要国家发文合作情况,从图2可以看出苏格兰、英格兰、澳大利亚、加拿大与其他国家合作较多。其中苏格兰与发文量较多的澳大利亚、英格兰、加拿大、法国、葡萄牙、丹麦均有合作。发文量最多的美国与主要研究国家中的日本有合作关系,日本还与澳大利亚和德国合作相对紧密,而中国仅与葡萄牙一个国家有过合作。
由表2可知,康奈尔大学、东北大学、京都大学发文量位居前三。康奈尔大学、东北大学、京都大学、奈良先端科学技术大学院大学、宾州州立大学这5个机构的全球引用总次数(total global citation score, TGCS)和本地引用总次数(total local citation score, TLCS)都较高。中国科学院和南京农业大学的发文量仅次于前5个机构,但是文章被引频次远远落后,在研究实力方面还存在一定差距,而2010年至今的发文量跃居前3,说明近年来在该领域加大了研究力度。伯明翰大学和墨尔本大学近6年的发文量均为零,表明这两个机构的研究重心可能发生了转移或者由基础研究转为应用研究。综合评价得出,康奈尔大学、东北大学、京都大学、奈良先端科学技术大学院大学和宾州州立大学是植物S位点自交不亲和领域研究的重要机构,中国科学院和南京农业大学也值得关注。
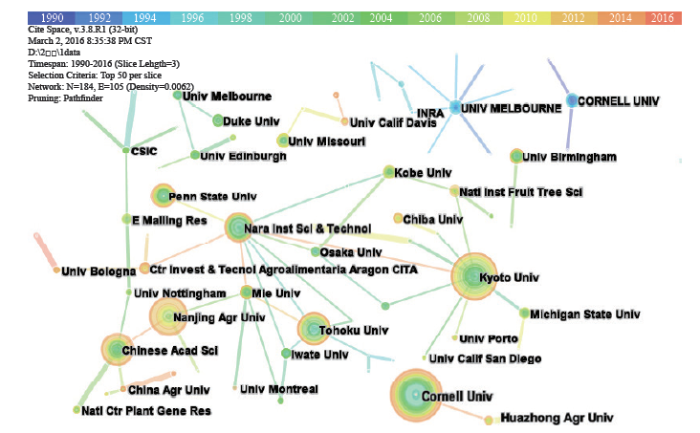
由图3可以看出,各机构间的合作不是十分密切,机构分布相对分散。较集中的合作网络有两个,分别是奈良先端科学技术大学院大学和京都大学。奈良先端科学技术大学院大学的合作对象都是该领域的主要研究机构,例如,东北大学、 宾州州立大学、三重大学、京都大学和岩手大学。发文量最多的康奈尔大学也仅与中国的华中农业大学合作较多。就中国的研究机构而言,中国科学院与南京农业大学和中国农业大学有合作,但合作也不够紧密。
对重要作者的分析是分析学科领域发展状况的重要环节,通过分析重要作者所发表的科研成果能对该学科领域的主要研究内容有大致了解。而分析作者分布及其合作网络是确定某研究领域核心作者群的一个重要方法,可以分析出作者对该研究领域的贡献度。本着发文量、全球总被引、本地总被引3项指标排行均靠前、同等条件下全球总被引指标优先的原则,选取了10位重要作者,并统计其作为通讯作者的发文量和近6年的发文量,结果见表3。其中ISOGAI A和HINATA K近6年的发文量为零,表明他们曾对该领域做出了巨大的贡献,但近几年的研究侧重点发生了改变,而其余8位作者一直持续植物自交不亲和机制的研究,值得关注。
从图4中可提取出与植物S位点自交不亲和机制研究相关的3个重要词:人工合成(resynthesized)、激酶(kinase)、磷酸化蛋白质组学(phosphoproteomic)。分析可知,植物S位点自交不亲和机制的研究可以追溯到1955年,自1990年之后,陆续涌现出大量研究成果。早期关于S基因的研究集中在人工合成的甘蓝型油菜上,近些年的研究热点为S位点受体激酶和蛋白质磷酸化。利用VosViewer进行主题词分析,可以得出研究前沿相关性较强的几个词:evolution、brassicaceae、arabidopsis,如图5中左侧红色区域所示,即十字花科,尤其是拟南芥的演化是其研究热点。结合图4、图5及文献解读可以得出该领域研究热点及前沿主要体现在以下3个方面:蛋白质磷酸化、S-RNase泛素化和SRK介导的拟南芥属的演化。
蛋白质磷酸化是生物体内广泛存在的一种调节方式,发生在SI信号转导的早期阶段,在调控柱头乳突细胞对细胞外的信号识别及细胞内的信号传递方面起着重要作用[15]。SRK的结构域通过胞外域特异性地与SCR结合,引起胞内激酶域的自磷酸化。M位点蛋白激酶(M locus protein kinase, MLPK)是甘蓝的一种非受体蛋白激酶,能够与SRK形成信号复合体。该信号复合体的磷酸化引起下游组分的级联反应,从而导致自交不亲和现象的发生[16]。Poulter等[10]也证实可溶性无机焦磷酸酶(soluble inorganic pyrophosphatases)和丝裂原活化蛋白激酶 (mitogen-activated protein kinases, MAPKs)的磷酸化是SI的下游反应。到目前为止,虽然已在SI信号转导通路中鉴定出了一些蛋白磷酸酶和蛋白激酶,但还是未能解释自交花粉受到抑制从而不亲和的原因,只有对整个信号通路蛋白质磷酸化途径有更加全面的认识,才能进一步阐明SI的分子机制[17]。前人对花粉与雌蕊相互作用的研究主要集中在用比较转录组学的方法来探讨基因表达的情况。然而,在蛋白质翻译后修饰的研究还较为缺乏。Wang等[18, 19]对水稻成熟雌蕊一个磷酸化蛋白质组学的剖析为研究植物自交不亲和的细胞信号转导提供了参考。
花柱特异性因子为S-RNase的SI在自然界中存在最为广泛,相应的花粉决定因子在车前科(Plantaginaceae)和茄科中被称为SLF(S-locus F-box),在蔷薇科中类似的F-box蛋白被命名为SFB(S haplotype-specific F-box protein)[20, 21]。Qiao等[22]通过免疫共沉淀和酵母双杂交实验发现并证明进入异己花粉管的S-RNase会与SLF结合并被泛素化。泛素蛋白酶体途径(ubiquitin proteasome pathway)介导的蛋白降解是机体调节细胞内蛋白水平与功能的重要机制,大量研究证实F-box是泛素蛋白降解系统中泛素蛋白连接酶复合体(SCF-E3)结合特异性底物的受体[23, 24]。针对SI系统,中科院遗传所薛勇彪课题组提出了一个S-RNase泛素化介导的液泡分拣模型[7]:S-RNase通过细胞内吞作用从细胞外基质进入到花粉管后被运输到花粉的液泡中。在自交的花粉管中,液泡膜由于某种机制会被破坏,S-RNase便被释放到细胞质中,引起细胞毒素效应从而抑制花粉管的生长;而在异交的花粉管中,液泡不被破坏,S-RNase不能发挥其细胞毒性,因此异交花粉管就可以正常生长。这个模型的提出,合理解释了花粉特征的特异性,为进一步研究植物自交不亲和反应的分子机制提供了重要基础。
S位点受体激酶是以十字花科为代表的SSI反应的核心因子。Stein等[25]首次从甘蓝中鉴定出柱头受体激酶SRK。Nasrallah JB[26]首先发现甘蓝自交不亲和特异性的花粉S基因SCR/SP11。在拟南芥中,SCR作为配体与受体SRK相互作用发挥自交不亲和效应。而甘蓝的自交不亲和性由SRK、SCR和S位点糖蛋白(SLG)3个高度多态性的基因共同控制,SLG不是必须的花柱S基因,但它的存在可以增强自交不亲和响应的能力[27, 28]。植物交配系统具有高度不稳定的特性,异交和自交之间频繁的转换,但是潜在的遗传原因还不明确。Mable等[29]基于温室条件下的授粉实验发现北美深山南芥中存在SI丢失的现象,并表明可能与SRK扩增的等位基因有关。Tsuchimatsu等[30]在欧洲拟南芥中发现了花粉特异性基因SCR中碱基的突变,进一步描述了拟南芥SI丢失的遗传机制。通过对拟南芥自交不亲和基因系统发育分析可知SCR与SRK存在共进化的关系[31]。拟南芥的祖先具有自交不亲和性,然而在漫长的适应环境的进化过程中逐渐向自交亲和过度。随着全基因组测序的发展,拟南芥属SI的丢失及某些基因的进化将会更加清晰全面的呈现出来。
本文运用计量学的手段对植物S位点自交不亲和机制研究领域进行了分析,我们可以得出,植物S位点自交不亲和机制领域受到越来越多学者的关注和研究。美国、日本、英格兰、加拿大在自交不亲和机制研究领域中具有较大的学术影响力,但本领域国际间的合作总体来说不够紧密,其中苏格兰、英格兰、澳大利亚、加拿大与其他国家间的合作相对较多。康奈尔大学、东北大学、京都大学、奈良先端科学技术大学院大学和宾州州立大学是植物S位点自交不亲和领域研究的重要机构。美国康奈尔大学的Nasrallah JB和Nasrallah ME、日本奈良先端科学技术大学院大学的Isogai A和Takayama S、日本东北大学的Watanabe M、Nishio T和Hinata K、美国宾州州立大学的Kao TH、日本京都大学的Tao R与日本千叶大学的Sassa H为该领域研究的前10位重要作者。蛋白质磷酸化、S-RNase泛素化和SRK介导的拟南芥属的演化是本领域近期或者未来几年的研究前沿热点。
植物自交不亲和反应可以发生在受精前的多个阶段,而且在自交不亲和系统中还存有一些重要的非S因子,与S因子存在着互作关系,体现出SI具有极其复杂的分子机制。近些年来,植物S位点自交不亲和机制的研究取得了一定进展,但仍然存在诸多问题:(1)蛋白质磷酸化与S-RNase泛素化有无互作关系,是如何相互作用的? (2)S-RNase进入自交的花粉管后,液泡膜是如何被破坏的?(3)S-RNase进入异交花粉管的细胞便不能发挥细胞毒素效应是否与花粉S因子有关?(4)其他自交不亲和植物中是否存在类似的花粉和花柱S因子?(5)现阶段普遍认为SI的机理是在花粉管或花粉中诱发了细胞程序性死亡,详细机制又是什么?要解决这一系列问题,笔者认为需要从以下3个层面进行深入研究:(1)在基因层面上,挖掘与SI相关的基因并进行单个甚至多个基因同时作用的功能分析。(2)在分子层面上,研究蛋白质体系、蛋白质-核酸体系是如何相互作用的。(3)在细胞层面上,探讨S-RNase进入液泡的信号转导途径及其他相关信号通路。同时需要加强各机构间的国际合作,运用分子遗传学、基因组学、生物信息学、细胞生物学、进化发育生物学等学科的综合手段对自交不亲和信号识别和激活的分子机制作进一步研究。不同种属自交不亲和性机制的阐明对人工打破其生殖障碍、开辟新的作物育种途径具有重要意义。随着生命科学的飞速发展,相信自交不亲和性的机制会在不远的将来得到详尽的诠释。
The authors have declared that no competing interests exist.
| [1] |
DOI:10.3969/j.issn.1000-6850.2012.18.030
URL
[Cite within: 1]
结合国内外最新的研究进展对配子体自交不亲和性的细胞学机制和分子生物学机制进行了综述,包括与S-核酸酶(S-RNase)、花粉管胞质自由钙离子以及其他因子相关的信号转导机制。并根据目前的研究现状提出了植物配子体自交不亲和机制研究中存在的问题及今后研究的重点,旨在为其深入研究提供参考。
|
| [2] |
DOI:10.1093/aob/mcq253
PMID:3170144
URL
[Cite within: 1]
BACKGROUND: For the Solanaceae-type self-incompatibility, also possessed by Rosaceae and Plantaginaceae, the specificity of self/non-self interactions between pollen and pistil is controlled by two polymorphic genes at the S-locus: the S-locus F-box gene (SLF or SFB) controls pollen specificity and the S-RNase gene controls pistil specificity. SCOPE: This review focuses on the work from the authors' laboratory using Petunia inflata (Solanaceae) as a model. Here, recent results on the identification and functional studies of S-RNase and SLF are summarized and a protein-degradation model is proposed to explain the biochemical mechanism for specific rejection of self-pollen tubes by the pistil. CONCLUSIONS: The protein-degradation model invokes specific degradation of non-self S-RNases in the pollen tube mediated by an SLF, and can explain compatible versus incompatible pollination and the phenomenon of competitive interaction, where SI breaks down in pollen carrying two different S-alleles. In Solanaceae, Plantaginaceae and subfamily Maloideae of Rosaceae, there also exist multiple S-locus-linked SLF/SFB-like genes that potentially function as the pollen S-gene. To date, only three such genes, all in P. inflata, have been examined, and they do not function as the pollen S-gene in the S-genotype backgrounds tested. Interestingly, subfamily Prunoideae of Rosaceae appears to possess only a single SLF/SFB gene, and competitive interaction, observed in Solanaceae, Plantaginaceae and subfamily Maloideae, has not been observed. Thus, although the cytotoxic function of S-RNase is an integral part of SI in Solanaceae, Plantaginaceae and Rosaceae, the function of SLF/SFB may have diverged. This highlights the complexity of the S-RNase-based SI mechanism. The review concludes by discussing some key experiments that will further advance our understanding of this self/non-self discrimination mechanism.
|
| [3] |
|
| [4] |
DOI:10.1007/s00425-006-0284-2
PMID:16794841
Magsci
URL
[Cite within: 1]
Self-incompatibility (SI) prevents the production of “self” seed and inbreeding by providing a recognition and rejection system for “self,” or genetically identical, pollen. Studies of gametophytic SI (GSI) species at a molecular level have identified two completely different S -genes and SI mechanisms. One GSI mechanism, which is found in the Solanaceae, Rosaceae and Scrophulariaceae, has S-RNase as the pistil S -component and an F-box protein as the pollen S -component. However, non- S -locus factors are also required. In an incompatible situation, the S-RNases degrade pollen RNA, thereby preventing pollen tube growth. Here, in the light of recent evidence, we examine alternative models for how compatible pollen escapes this cytotoxic activity. The other GSI mechanism, so far found only in the Papaveraceae, has a small secreted peptide, the S-protein, as its pistil S -component. The pollen S -component remains elusive, but it is thought to be a transmembrane receptor, as interaction of the S-protein with incompatible pollen triggers a signaling network, resulting in rapid actin depolymerization and pollen tube inhibition and programmed cell death (PCD). Here, we present an overview of what is currently known about the mechanisms involved in regulating pollen tube inhibition in these two GSI systems.
|
| [5] |
DOI:10.1007/s00497-009-0111-6
PMID:20165962
Magsci
URL
[Cite within: 1]
Many species of Rosaceae, Solanaceae, and Plantaginaceae exhibit S-RNase-based self-incompatibility (SI) in which pistil-part specificity is controlled by S locus-encoded ribonuclease (S-RNase). Although recent findings revealed that S locus-encoded F-box protein, SLF/SFB, determines pollen-part specificity, how these pistil- and pollen-part S locus products interact in vivo and elicit the SI reaction is largely unclear. Furthermore, genetic studies suggested that pollen S function can differ among species. In Solanaceae and the rosaceous subfamily Maloideae (e.g., apple and pear), the coexistence of two different pollen S alleles in a pollen breaks down SI of the pollen, a phenomenon known as competitive interaction. However, competitive interaction seems not to occur in the subfamily Prunoideae (e.g., cherry and almond) of Rosaceae. Furthermore, the effect of the deletion of pollen S seems to vary among taxa. This review focuses on the potential differences in pollen-part function between subfamilies of Rosaceae, Maloideae, and Prunoideae, and discusses implications for the mechanistic divergence of the S-RNase-based SI.
|
| [6] |
DOI:10.1016/j.tplants.2003.10.007
PMID:14659710
URL
[Cite within: 1]
Many hermaphrodite flowering plants avoid self-fertilization through genetic systems of self-incompatibility (SI). SI allows a plant to recognize and to reject self or self-related pollen, thereby preserving its ovules for outcrossing. Genes situated at the S -locus encode the ‘male’ (pollen) and ‘female’ (pistil) recognition determinants of SI. In sporophytic SI (SSI) the male determinant is expressed in the diploid anther, therefore haploid pollen grains behave with a diploid S phenotype. In Brassica , the male and the female determinants of SSI have been identified as a peptide ligand and its cognate receptor, respectively, and recent studies have identified downstream signalling molecules involved in pollen rejection. It now needs to be established whether the Brassica mechanism is universal in species with SSI, or unique to the Brassicaceae.
|
| [7] |
|
| [8] |
DOI:10.1093/molbev/msl042
PMID:16782760
URL
[Cite within: 1]
A recent investigation found evidence that the transition of Arabidopsis thaliana from ancestral self-incompatibility (SI) to full self-compatibility occurred very recently and suggested that this occurred through a selective fixation of a nonfunctional allele (WSCR1) at the SCR gene, which determines pollen specificity in the incompatibility response. The main evidence is the lack of polymorphism at the SCR locus in A. thaliana. However, the nearby SRK gene, which determines stigma specificity in self-incompatible Brassicaceae species, has extremely high sequence diversity, with 3 very divergent SRK haplotypes, 2 of them present inmultiple strains. Such high diversity is extremely unusual in this species, and it suggests the possibility that multiple, different SRK haplotypes may have been preserved fromA. thaliana's self-incompatible ancestor. To study the evolution of S-haplotypes in the A. thaliana lineage, we searched the 2 most closely related Arabidopsis species Arabidopsis lyrata and Arabidopsis halleri, in which most populations have retained SI, and found SRK sequences corresponding to all 3 A. thaliana haplogroup sequences. Our molecular evolutionary analyses of these 3 S-haplotypes provide an independent estimate of the timing of the breakdown of SI and again exclude an ancient transition to selfing in A. thaliana. Comparing sequences of each of the 3 haplogroups between species, we find that 2 of the 3 SRK sequences (haplogroups A and B) are similar throughout their length, suggesting that little or no recombination with other SRK alleles has occurred since these species diverged. The diversity difference between the SCR and SRK loci in A. thaliana, however, suggests crossingover, either within SRK or between the SCR and SRK loci. If the loss of SI involved fixation of the WSCR1 sequence, the exchange must have occurred during its fixation. Divergence between the species is much lower at the S-locus, compared with reference loci, andwe discuss two contributory possibilities. Introgression may have occurred between A. lyrata and A. halleri and between their ancestral lineage and A. thaliana, at least for some period after their split. In addition, the coalescence times of sequences of individual S-haplogroups are expected to be less than those of alleles at non鈥揝-loci.
|
| [9] |
DOI:10.2307/3871411
PMID:11251101
URL
[Cite within: 2]
Abstract As a starting point for a phylogenetic study of self-incompatibility (SI) in crucifers and to elucidate the genetic basis of transitions between outcrossing and self-fertilizing mating systems in this family, we investigated the SI system of Arabidopsis lyrata. A. lyrata is an outcrossing close relative of the self-fertile A. thaliana and is thought to have diverged from A. thaliana approximately 5 million years ago and from Brassica spp 15 to 20 million years ago. Analysis of two S (sterility) locus haplotypes demonstrates that the A. lyrata S locus contains tightly linked orthologs of the S locus receptor kinase (SRK) gene and the S locus cysteine-rich protein (SCR) gene, which are the determinants of SI specificity in stigma and pollen, respectively, but lacks an S locus glycoprotein gene. As described previously in Brassica, the S haplotypes of A. lyrata differ by the rearranged order of their genes and by their variable physical sizes. Comparative mapping of the A. lyrata and Brassica S loci indicates that the S locus of crucifers is a dynamic locus that has undergone several duplication events since the Arabidopsis--Brassica split and was translocated as a unit between two distant chromosomal locations during diversification of the two taxa. Furthermore, comparative analysis of the S locus region of A. lyrata and its homeolog in self-fertile A. thaliana identified orthologs of the SRK and SCR genes and demonstrated that self-compatibility in this species is associated with inactivation of SI specificity genes.
|
| [10] |
DOI:10.1042/BST0380588
PMID:20298226
URL
[Cite within: 2]
Many flowering plants are hermaphrodite, posing the problem of self-fertilization and the subsequent loss of the genetic fitness of the offspring. To prevent this, many plants have developed a genetically controlled mechanism called self-incompatibility (SI). When the male and female S-determinants match, self (incompatible) pollen is recognized and rejected before fertilization can occur. In poppy (Papaver rhoeas), the pistil S-determinant (PrsS) is a small secreted protein that interacts with incompatible pollen, initiating a Ca(2+)-dependent signalling network. SI triggers several downstream events, including depolymerization of the cytoskeleton, phosphorylation of two soluble inorganic pyrophosphatases and an MAPK (mitogen-activated protein kinase). This culminates in PCD (programmed cell death) involving several caspase-like activities. The recent discovery of the Papaver pollen S-determinant PrpS marks a significant step forward in the understanding of the Papaver SI system. PrpS encodes a ~20 kDa predicted transmembrane protein which has no homology with known proteins. It is specifically expressed in pollen, linked to the pistil S-determinant, and displays the high polymorphism expected of an S-locus determinant. The present review focuses on the discovery and characterization of PrpS which strongly support the hypothesis that Papaver SI is triggered by the interaction of PrsS and PrpS.
|
| [11] |
DOI:10.1534/genetics.104.031146
PMID:15371361
URL
[Cite within: 1]
As a model system in classical plant genetics, the genus Antirrhinum has been well studied, especially in gametophytic self-incompatibility, flower development biology, and transposon-induced mutation. In contrast to the advances in genetic and molecular studies, little is known about Antirrhinum cytogenetics. In this study, we isolated two tandem repetitive sequences, CentA1 and CentA2, from the centromeric regions of Antirrhinum chromosomes. A standard karyotype has been established by anchoring these centromeric repeats on meiotic pachytene chromosome using FISH. An ideogram based on the DAPI-staining pattern of pachytene chromosomes was developed to depict the distribution of heterochromatin in the Antirrhinum majus genome. To integrate the genetic and chromosomal maps, we selected one or two molecular markers from each linkage group to screen an Antirrhinum transformation-competent artificial chromosome (TAC) library. These genetically anchored TAC clones were labeled as FISH probes to hybridize to pachytene chromosomes of A. majus. As a result, the relationship between chromosomes and the linkage groups (LGs) in Antirrhinum has been established.
|
| [12] |
DOI:10.1006/anbo.1999.1056
URL
[Cite within: 1]
The grasses represent one of the most important families of plants since they constitute our major crops and pastures and dominate many natural ecosystems. Self-incompatibility (SI) is widespread in the grasses and is under the control of a series of alleles at each of two unlinked loci, S and Z . Specification of the pollen grain is gametophytic and depends upon the combination of S and Z alleles in the pollen grain. Both of these must be matched in the pistil for the pollen grain to be incompatible. Several lines of research suggest that this two-locus SI system is shared by all grass species of the Pooideae and possibly by the entire Graminae. Firstly, genetic studies demonstrated the presence of the S 鈥 Z system in the Triticeae, the Poeae and Avenae. Secondly, linkage analyses showed common markers linked to the S -gene in grass species from all three tribes. Recent molecular studies revealed remarkable synteny of chromosomes and conservation of gene order within the Poaceae. Molecular markers have also been used to confirm syntenous localization of S and Z in several species. Despite the complexity of the grass incompatibility system relative to that in other families, the detailed genetic and physical maps for the crop species in the Triticeae and the availability of large insert libraries and mutations at the incompatibility loci, makes this a particularly promising family for molecular studies.
|
| [13] |
|
| [14] |
DOI:10.1093/jxb/erp383
PMID:20097844
URL
[Cite within: 1]
Cell-cell communication is vital to multicellular organisms and much of it is controlled by the interactions of secreted protein ligands (or other molecules) with cell surface receptors. In plants, receptor-ligand interactions are known to control phenomena as diverse as floral abscission, shoot apical meristem maintenance, wound response, and self-incompatibility (SI). SI, in which 'self' (incompatible) pollen is rejected, is a classic cell-cell recognition system. Genetic control of SI is maintained by an S-locus, in which male (pollen) and female (pistil) S-determinants are encoded. In Papaver rhoeas, PrsS proteins encoded by the pistil S-determinant interact with incompatible pollen to effect inhibition of pollen growth via a Ca(2+)-dependent signalling network, resulting in programmed cell death of 'self' pollen. Recent studies are described here that identified and characterized the pollen S-determinant of SI in P. rhoeas. Cloning of three alleles of a highly polymorphic pollen-expressed gene, PrpS, which is linked to pistil-expressed PrsS revealed that PrpS encodes a novel approximately 20 kDa transmembrane protein. Use of antisense oligodeoxynucleotides provided data showing that PrpS functions in SI and is the pollen S-determinant. Identification of PrpS represents a milestone in the SI field. The nature of PrpS suggests that it belongs to a novel class of 'receptor' proteins. This opens up new questions about plant 'receptor'-ligand pairs, and PrpS-PrsS have been examined in the light of what is known about other receptors and their protein-ligand pairs in plants.
|
| [15] |
DOI:10.1146/annurev.cellbio.16.1.333
PMID:11031240
URL
[Cite within: 1]
Many bisexual flowering plants possess a reproductive strategy called self-incompatibility (SI) that enables the female tissue (the pistil) to reject self but accept non-self pollen for fertilization. Three different SI mechanisms are discussed, each controlled by two separate, highly polymorphic genes at the S-locus. For the Solanaceae and Papaveraceae types, the genes controlling female function in SI, the S-RNase gene and the S-gene, respectively, have been identified. For the Brassicaceae type, the gene controlling male function, SCR/SP11, and the gene controlling female function, SRK, have been identified. The S-RNase based mechanism involves degradation of RNA of self-pollen tubes; the S-protein based mechanism involves a signal transduction cascade in pollen, including a transient rise in [Ca(2+)]i and subsequent protein phosphorylation/dephosphorylation; and the SRK (a receptor kinase) based mechanism involves interaction of a pollen ligand, SCR/SP11, with SRK, followed by a signal transduction cascade in the stigmatic surface cell.
|
| [16] |
DOI:10.1126/science.1093586
PMID:15001779
URL
[Cite within: 1]
Self-incompatibility (SI) response in Brassica is initiated by haplotype-specific interactions between the pollen-borne ligand S locus protein 11/SCR and its stigmatic S receptor kinase, SRK. This binding induces autophosphorylation of SRK, which is then thought to trigger a signaling cascade that leads to self-pollen rejection. A recessive mutation of the modifier (m) gene eliminates the SI response in stigma. Positional cloning of M has revealed that it encodes a membrane-anchored cytoplasmic serine/threonine protein kinase, designated M locus protein kinase (MLPK). Transient expression of MLPK restores the ability of mm papilla cells to reject self-pollen, suggesting that MLPK is a positive mediator of Brassica SI signaling.
|
| [17] |
DOI:10.1371/journal.pone.0140507
PMID:4616620
URL
[Cite within: 1]
Xanthoceras sorbifolium, a tree species endemic to northern China, has high oil content in its seeds and is recognized as an important biodiesel crop. The plant is characterized by late-acting self-incompatibility (LSI). LSI was found to occur in many angiosperm species and plays an important role in reducing inbreeding and its harmful effects, as do gametophytic self-incompatibility (GSI) and sporophytic self-incompatibility (SSI). Molecular mechanisms of conventional GSI and SSI have been well characterized in several families, but no effort has been made to identify the genes involved in the LSI process. The present studies indicated that there were no significant differences in structural and histological features between the self- and cross-pollinated ovules during the early stages of ovule development until 5 days after pollination (DAP). This suggests that 5 DAP is likely to be a turning point for the development of the selfed ovules. Comparative de novo transcriptome analysis of the selfed and crossed ovules at 5 DAP identified 274 genes expressed specifically or preferentially in the selfed ovules. These genes contained a significant proportion of genes predicted to function in the biosynthesis of secondary metabolites, consistent with our histological observations in the fertilized ovules. The genes encoding signal transduction-related components, such as protein kinases and protein phosphatases, are overrepresented in the selfed ovules. X. sorbifolium selfed ovules also specifically or preferentially express many unique transcription factor (TF) genes that could potentially be involved in the novel mechanisms of LSI. We also identified 42 genes significantly up-regulated in the crossed ovules compared to the selfed ovules. The expression of all 16 genes selected from the RNA-seq data was validated using PCR in the selfed and crossed ovules. This study represents the first genome-wide identification of genes expressed in the fertilized ovules of an LSI species. The availability of a pool of specifically or preferentially expressed genes from selfed ovules for X. sorbifolium will be a valuable resource for future genetic analyses of candidate genes involved in the LSI response.
|
| [18] |
DOI:10.1002/pmic.201400004
PMID:25074045
Magsci
URL
[Cite within: 1]
As the female reproductive part of a flower, the pistil consists of the ovary, style, and stigma, and is a critical organ for the process from pollen recognition to fertilization and seed formation. Previous studies on pollen-pistil interaction mainly focused on gene expression changes with comparative transcriptomics or proteomics method. However, studies on protein PTMs are still lacking. Here we report a phosphoproteomic study on mature pistil of rice. Using IMAC enrichment, hydrophilic interaction chromatography fraction and high-accuracy MS instrument (TripleTOF 5600), 2347 of high-confidence (Ascore 鈮 19, p 鈮 0.01), phosphorylation sites corresponding to 1588 phosphoproteins were identified. Among them, 1369 phosphorylation sites within 654 phosphoproteins were newly identified; 41 serine phosphorylation motifs, which belong to three groups: proline-directed, basophilic, and acidic motifs were identified after analysis by motif-X. Two hundred and one genes whose phosphopeptides were identified here showed tissue-specific expression in pistil based on information mining of previous microarray data. All MS data have been deposited in the ProteomeXchange with identifier PXD000923 (http://proteomecentral.proteomexchange.org/dataset/PXD000923). This study will help us to understand pistil development and pollination on the posttranslational level.
|
| [19] |
DOI:10.1007/s11103-015-0410-2
PMID:26613898
URL
[Cite within: 1]
Rice (Oryza sativa L.) seed serves as a major food source for over half of the global population. Though it has been long recognized that phosphorylation plays an essential role in rice seed development, the phosphorylation events and dynamics in this process remain largely unknown so far. Here, we report the first large scale identification of rice seed phosphoproteins and phosphosites by using a quantitative phosphoproteomic approach. Thorough proteomic studies in pistils and seeds at 3, 7聽days after pollination resulted in the successful identification of 3885, 4313 and 4135 phosphopeptides respectively. A total of 2487 proteins were differentially phosphorylated among the three stages, including Kip related protein 1, Rice basic leucine zipper factor 1, Rice prolamin box binding factor and numerous other master regulators of rice seed development. Moreover, differentially phosphorylated proteins may be extensively involved in the biosynthesis and signaling pathways of phytohormones such as auxin, gibberellin, abscisic acid and brassinosteroid. Our results strongly indicated that protein phosphorylation is a key mechanism regulating cell proliferation and enlargement, phytohormone biosynthesis and signaling, grain filling and grain quality during rice seed development. Overall, the current study enhanced our understanding of the rice phosphoproteome and shed novel insight into the regulatory mechanism of rice seed development.
|
| [20] |
DOI:10.1126/science.1195243
PMID:21051632
URL
[Cite within: 1]
Self-incompatibility in flowering plants prevents inbreeding and promotes outcrossing to generate genetic diversity. In Solanaceae, a multiallelic gene, S-locus F-box (SLF), was previously shown to encode the pollen determinant in self-incompatibility. It was postulated that an SLF allelic product specifically detoxifies its non-self S-ribonucleases (S-RNases), allelic products of the pistil determinant, inside pollen tubes via the ubiquitin-26S-proteasome system, thereby allowing compatible pollinations. However, it remained puzzling how SLF, with much lower allelic sequence diversity than S-RNase, might have the capacity to recognize a large repertoire of non-self S-RNases. We used in vivo functional assays and protein interaction assays to show that in Petunia, at least three types of divergent SLF proteins function as the pollen determinant, each recognizing a subset of non-self S-RNases. Our findings reveal a collaborative non-self recognition system in plants.
|
| [21] |
DOI:10.1016/j.envsoft.2012.05.002
PMID:12615948
URL
[Cite within: 1]
Abstract Gametophytic self-incompatibility in Rosaceae, Solanaceae, and Scrophulariaceae is controlled by the S locus, which consists of an S-RNase gene and an unidentified "pollen S" gene. An approximately 70-kb segment of the S locus of the rosaceous species almond, the S haplotype-specific region containing the S-RNase gene, was sequenced completely. This region was found to contain two pollen-expressed F-box genes that are likely candidates for pollen S genes. One of them, named SFB (S haplotype-specific F-box protein), was expressed specifically in pollen and showed a high level of S haplotype-specific sequence polymorphism, comparable to that of the S-RNases. The other is unlikely to determine the S specificity of pollen because it showed little allelic sequence polymorphism and was expressed also in pistil. Three other S haplotypes were cloned, and the pollen-expressed genes were physically mapped. In all four cases, SFBs were linked physically to the S-RNase genes and were located at the S haplotype-specific region, where recombination is believed to be suppressed, suggesting that the two genes are inherited as a unit. These features are consistent with the hypothesis that SFB is the pollen S gene. This hypothesis predicts the involvement of the ubiquitin/26S proteasome proteolytic pathway in the RNase-based gametophytic self-incompatibility system.
|
| [22] |
|
| [23] |
DOI:10.1105/tpc.114.126920
PMID:25070642
Magsci
URL
[Cite within: 1]
Petunia possesses self-incompatibility, by which pistils reject self-pollen but accept non-self-pollen for fertilization. Self-/non-self-recognition between pollen and pistil is regulated by the pistil-specific S-RNase gene and by multiple pollen-specific S-locus F-box (SLF) genes. To date, 10 SLF genes have been identified by various methods, and seven have been shown to be involved in pollen specificity. For a given S-haplotype, each SLF interacts with a subset of its non-self S-RNases, and an as yet unknown number of SLFs are thought to collectively mediate ubiquitination and degradation of all non-self S-RNases to allow cross-compatible pollination. To identify a complete suite of SLF genes of P. inflata, we used a de novo RNA-seq approach to analyze the pollen transcriptomes of S2-haplotype and S3-haplotype, as well as the leaf transcriptome of the S3S3 genotype. We searched for genes that fit several criteria established from the properties of the known SLF genes and identified the same seven new SLF genes in S2-haplotype and S3-haplotype, suggesting that a total of 17 SLF genes constitute pollen specificity in each S-haplotype. This finding lays the foundation for understanding how multiple SLF genes evolved and the biochemical basis for differential interactions between SLF proteins and S-RNases.
|
| [24] |
DOI:10.1371/journal.pone.0097642
PMID:4029751
URL
[Cite within: 1]
Gametophytic self-incompatibility (GSI) of Rosaceae, Solanaceae and Plantaginaceae is controlled by a single polymorphic S locus. The S locus contains at least two genes, S-RNase and F-box protein encoding gene SLF/SFB/SFBB that control pistil and pollen specificity, respectively. Generally, the F-box protein forms an E3 ligase complex, SCF complex with Skp1, Cullin1 (CUL1) and Rbx1, however, in Petunia inflata, SBP1 (S-RNase binding protein1) was reported to play the role of Skp1 and Rbx1, and form an SCFSLF-like complex for ubiquitination of non-self S-RNases. On the other hand, in Petunia hybrida and Petunia inflata of Solanaceae, Prunus avium and Pyrus bretschneideri of Rosaceae, SSK1 (SLF-interacting Skp1-like protein1) is considered to form the SCFSLF/SFB complex. Here, we isolated pollen-expressed apple homologs of SSK1 and CUL1, and named MdSSK1, MdCUL1A and MdCUL1B. MdSSK1 was preferentially expressed in pollen, but weakly in other organs analyzed, while, MdCUL1A and MdCUL1B were almost equally expressed in all the organs analyzed. MdSSK1 transcript abundance was significantly (>100 times) higher than that of MdSBP1. In vitro binding assays showed that MdSSK1 and MdSBP1 interacted with MdSFBB1-S9 and MdCUL1, and MdSFBB1-S9 interacted more strongly with MdSSK1 than with MdSBP1. The results suggest that both MdSSK1-containing SCFSFBB1 and MdSBP1-containing SCFSFBB1-like complexes function in pollen of apple, and the former plays a major role.
|
| [25] |
DOI:10.1073/pnas.88.19.8816
PMID:1681543
URL
[Cite within: 1]
Self-recognition between pollen and stigma during pollination in Brassica oleracea is genetically controlled by the multiallelic self-incompatibility locus (S). The authors describe the S receptor kinase (SRK) gene, a previously uncharacterized gene that residues at the S locus. The nucleotide sequences of genomic DNA and of cDNAs corresponding to SRK predict a putative transmembrane receptor having serine/threonine-specific protein kinase activity. Its extracellular domain exhibits striking homology to the secreted product of the S-locus genotypes are highly polymorphic and have apparently evolved in unison with genetically linked alleles of SLG. SRK directs the synthesis of several alternative transcripts, which potentially encode different protein products, and these transcripts were detected exclusively in reproductive organs. The identification of SRK may provide new perspectives into the signal transduction mechanism underlying pollen recognition.
|
| [26] |
DOI:10.1126/science.286.5445.1697
PMID:10576728
URL
[Cite within: 1]
Abstract In the S locus-controlled self-incompatibility system of Brassica, recognition of self-related pollen at the surface of stigma epidermal cells leads to inhibition of pollen tube development. The female (stigmatic) determinant of this recognition reaction is a polymorphic transmembrane receptor protein kinase encoded at the S locus. Another highly polymorphic, anther-expressed gene, SCR, also encoded at the S locus, fulfills the requirements for the hypothesized pollen determinant. Loss-of-function and gain-of-function studies prove that the SCR gene product is necessary and sufficient for determining pollen self-incompatibility specificity, possibly by acting as a ligand for the stigmatic receptor.
|
| [27] |
|
| [28] |
DOI:10.1038/35002628
PMID:10706292
URL
[Cite within: 1]
The self-incompatibility possessed by Brassica is an intraspecific reproductive barrier by which the stigma rejects self-pollen but accepts non-self-pollen for fertilization. The molecular/biochemical bases of recognition and rejection have been intensively studied. Self-incompatibility in Brassica is sporophytically controlled by the polymorphic S locus. Two tightly linked polymorphic genes at the S locus, S receptor kinase gene (SRK) and S locus glycoprotein gene (SLG), are specifically expressed in the papillar cells of the stigma, and analyses of self-compatible lines of Brassica have suggested that together they control stigma function in self-incompatibility interactions. Here we show, by transforming self-incompatible plants of Brassica rapa with an SRK28 and an SLG28 transgene separately, that expression of SRK28 alone, but not SLG28 alone, conferred the ability to reject self (S28)-pollen on the transgenic plants. We also show that the ability of SRK28 to reject S28 pollen was enhanced by SLG28. We conclude that SRK alone determines S haplotype specificity of the stigma, and that SLG acts to promote a full manifestation of the self-incompatibility response.
|
| [29] |
DOI:10.1111/j.0014-3820.2005.tb01794.x
PMID:16153030
URL
[Cite within: 1]
Abstract Mating systems in plants are known to be highly labile traits, with frequent transitions from outcrossing to selfing. The genetic basis for breakdown in self-incompatibility (SI) systems has been studied, but data on variation in selfing rates in species for which the molecular basis of SI is known are rare. This study surveyed such variation in Arabidopsis lyrata (Brassicaceae), which is often considered an obligately outcrossing species, to examine the causes and genetic consequences of changes in its breeding system. Based on controlled self-pollinations in the greenhouse, three populations from the Great Lakes region of North America included a minority of self-compatible (SC) individuals, while two showed larger proportions of SC individuals and all populations contained some individuals capable of setting selfed seeds. Loss of SI was not associated with particular haplotypes at the S-locus (as estimated by alleles amplified at the SRK locus, the gene controlling female specificity) and all populations contained similar numbers of SRK alleles, suggesting that some other genetic factor is responsible for modifying the SI reaction. The loss of SI has resulted in an effective shift in the mating system, as the two populations with a high frequency of SC individuals showed significantly lower microsatellite-based multilocus outcrossing rates and higher inbreeding coefficients than the other populations. Based on microsatellites, observed heterozygosities and genetic diversity were also significantly depressed in these populations. These findings provide the unique opportunity to examine in detail the consequences of mating system changes within a species with a well-characterized SI system.
|
| [30] |
DOI:10.1038/nature08927
PMID:20400945
URL
[Cite within: 1]
Ever since Darwin's pioneering research, the evolution of self-fertilisation (selfing) has been regarded as one of the most prevalent evolutionary transitions in flowering plants. A major mechanism to prevent selfing is the self-incompatibility (SI) recognition system, which consists of male and female specificity genes at the S-locus and SI modifier genes. Under conditions that favour selfing, mutations disabling the male recognition component are predicted to enjoy a relative advantage over those disabling the female component, because male mutations would increase through both pollen and seeds whereas female mutations would increase only through seeds. Despite many studies on the genetic basis of loss of SI in the predominantly selfing plant Arabidopsis thaliana, it remains unknown whether selfing arose through mutations in the female specificity gene (S-receptor kinase, SRK), male specificity gene (S-locus cysteine-rich protein, SCR; also known as S-locus protein 11, SP11) or modifier genes, and whether any of them rose to high frequency across large geographic regions. Here we report that a disruptive 213-base-pair (bp) inversion in the SCR gene (or its derivative haplotypes with deletions encompassing the entire SCR-A and a large portion of SRK-A) is found in 95% of European accessions, which contrasts with the genome-wide pattern of polymorphism in European A. thaliana. Importantly, interspecific crossings using Arabidopsis halleri as a pollen donor reveal that some A. thaliana accessions, including Wei-1, retain the female SI reaction, suggesting that all female components including SRK are still functional. Moreover, when the 213-bp inversion in SCR was inverted and expressed in transgenic Wei-1 plants, the functional SCR restored the SI reaction. The inversion within SCR is the first mutation disrupting SI shown to be nearly fixed in geographically wide samples, and its prevalence is consistent with theoretical predictions regarding the evolutionary advantage of mutations in male components.
|
| [31] |
DOI:10.1534/g3.114.010843
PMID:24939184
URL
[Cite within: 1]
Self-incompatibility (SI) is a genetic system that prevents self-fertilization in many Angiosperms. Although plants from the Brassicaceae family present an apparently unique SI system that is ancestral to the family, investigations at the S-locus responsible for SI have been mostly limited to two distinct lineages (Brassica and Arabidopsis-Capsella, respectively). Here, we investigated SI in a third deep-branching lineage of Brassicaceae: the tribe Biscutelleae. By coupling sequencing of the SI gene responsible for pollen recognition (SRK) with phenotypic analyses based on controlled pollinations, we identified 20 SRK-like sequences functionally linked to 13 S-haplotypes in 21 individuals of Biscutella neustriaca and 220 seedlings. We found two genetic and phylogenetic features of SI in Biscutelleae that depart from patterns observed in the reference Arabidopsis clade: (1) SRK-like sequences cluster into two main phylogenetic lineages interspersed within the many SRK lineages of Arabidopsis; and (2) some SRK-like sequences are transmitted by linked pairs, suggesting local duplication within the S-locus. Strikingly, these features also were observed in the Brassica clade but probably evolved independently, as the two main SRK clusters in Biscutella are distinct from those in Brassica. In the light of our results and of what has been previously observed in other Brassicaceae, we discuss the ecological and evolutionary implications on SI plant populations of the high diversity and the complex dominance relationships we found at the S-locus in Biscutelleae.
|